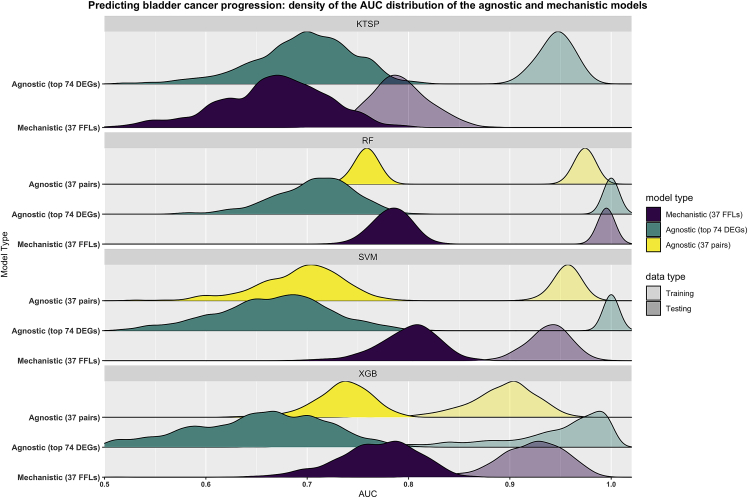
Figure 2.
Mechanistic models based on FFLs outperform agnostic ones in predicting bladder cancer progression
The figure depicts the performance of the agnostic and mechanistic models as obtained using the described bootstrap design. Briefly, all models were trained on 1000 bootstraps of the training data (transparent colors), then evaluated on untouched testing data (solid colors) using the Area Under the ROC Curve (AUC) as performance metric. Mechanistic models were based on the feedforward loops (37 pairs) (purple) and agnostic models were trained either using the top differentially expressed genes (74 genes) (green) or the corresponding pairwise comparisons (37 pairs) (yellow). Curves represent the smoothed density distributions of the AUC values, and each panel corresponds to one of the four algorithms used. KTSP: K-top scoring pairs; RF: random forest; SVM: support vector machine; XGB: extreme gradient boosting; FFLs: feedforward loops; DEGs: differentially expressed genes.