Figure 5.
Validation of reporter OPCs generated using an independent, male hESC line (RUES1)
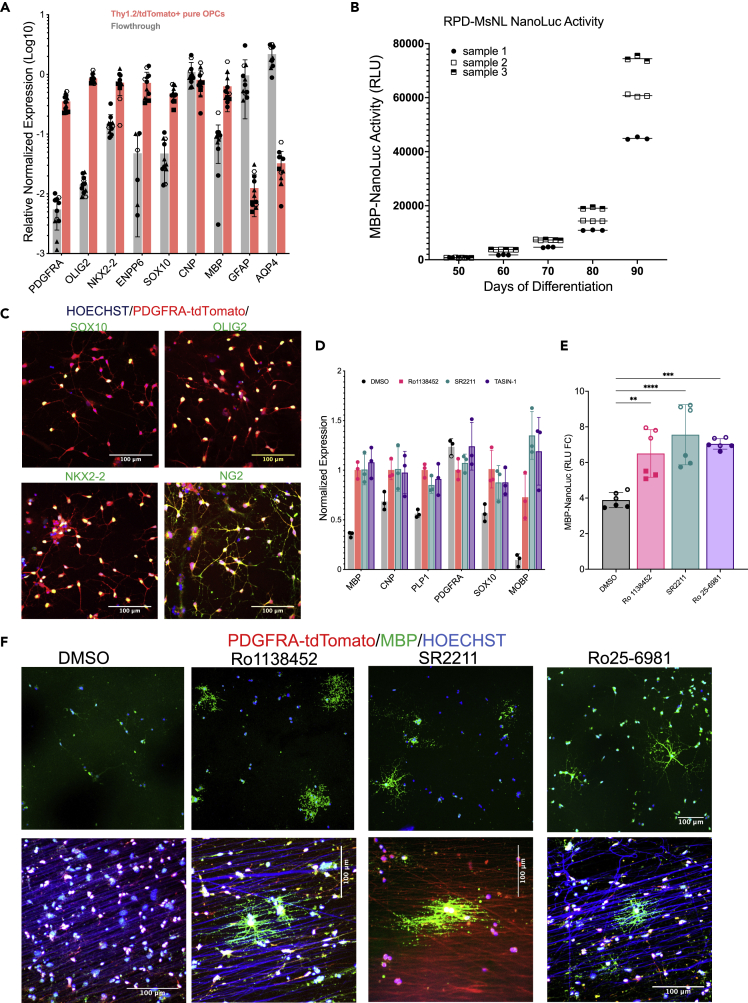
(A) Differentiated OPCs were MACS purified for the expression of PDGFRA-tdTomato-thy1.2, and the expression of different OPC markers between the tdTomato+ and tdTomato- (flow through) population were quantified by qPCR analysis, which shows enrichment of OPC markers in the tdTomato enriched population compared to the flow through. Three biological and three technical replicates each were used for qPCR analysis. Data are presented as mean SD. Biological and technical replicates are distinguished by filled vs clear symbols used for each data point. Source data for the qPCR are provided as a Source Data file.
(B) secNluc activity in the cell culture medium of differentiating RPD-MsNL reporter line measured with Nano-Glo assay. 20 μL of culture media from different days of differentiation (x-axis) were removed for the assay. Increase in Nano-Luc activity (y-axis), which corresponds to MBP expression increases as cells mature.
(C) Immunohistochemistry demonstrating that the MACS purified tdTomato+ cells express the OPC markers SOX10, OLIG2, NKX2.2 and NG2. Immunohistochemistry was independently repeated three times with similar results.
(D-F) qPCR (D), NanoLuc expression (E), and immunostaining (F) of the purified RPD-MsNL OPCs treated with different compounds for 10 days. (E) NanoLuc data represents two different experiments performed using different number of OPCs, represented as open (10K cells per treatment) versus filled (20k cells per treatment) symbol. (F) Stronger MBP staining, more MBP+ cells (top panel) and better myelination of the electrospun nanofibers ((950 nM diameter nanofibers) lower panel) by the compound treated OPCs compared to the DMSO treated cells is noticeable. Red channel was overexposed to visualize the nanofibers in the lower panel. Immunostaining was independently repeated three times with similar results. Data are presented as mean ± SD, scale bar: 100 µm.