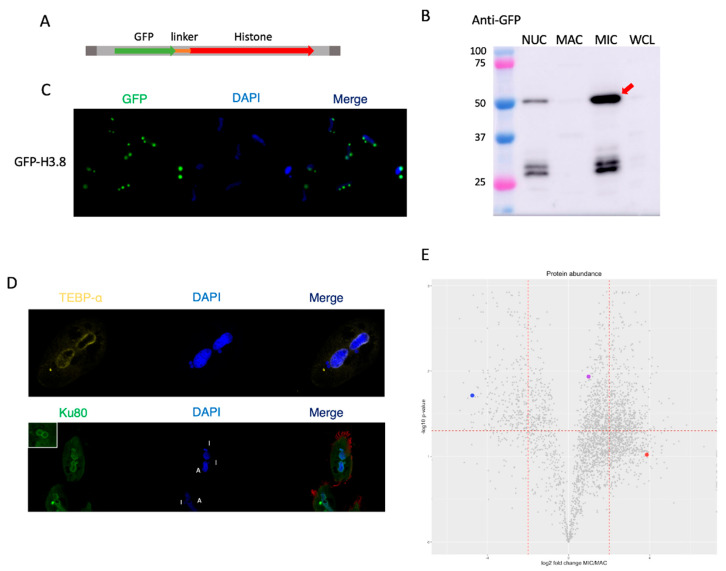
Figure 5.
Validation of nuclear localization results from MS. (A) Schematic of the artificial chromosome used to transform Oxytricha with an N-terminal GFP-tagged H3 histone. A ciliate codon-corrected GFP was inserted at the N-terminus of the gene along with a flexible linker. The double-stranded portion of the nanochromosome’s telomeres were included at both ends. (B) Western blot analysis of samples from cells transformed with the previously described construct including unseparated nuclei (NUC), enriched MAC, enriched MIC, and whole cell lysate (WCL) using an anti-GFP antibody. The expected band of approximately 48 kDa is marked with an arrow. The smaller bands between 25 and 37 kDa are close to the size of free GFP (27 kDa). (C) Fluorescence imaging of the GFP-tagged strain depicting the specific MIC localization of the transgene with DAPI to visualize the nuclei. (D) Immunofluorescence imaging of vegetative JRB310 cells using custom antibodies raised against Oxytricha TEBP-α or Ku80, with DAPI to visualize nuclei demonstrating the MAC-specific localization of TEBP-α and the nonspecific nuclear localization of Ku80. (E) Volcano plot with the 3 validated proteins depicted as colored spots. TEBP-α is labeled in blue, Ku80 in purple, and H3.8 in red.