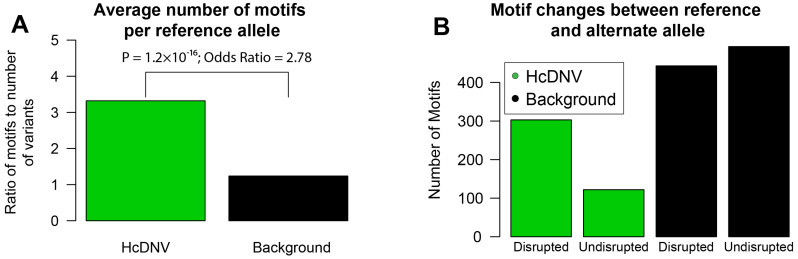
Figure 4.
Transcription factor binding motif disruption in HcDNV variants. Fimo was used to call transcription factor binding motifs that overlapped with the variant position. Fisher’s exact test was used to find the p-value and odds ratio for the proportions in (A,B). (A) Barplot showing the ratio of the number of filtered motifs found over the number of variant sequences used to find overlapping motifs. (B) Barplot showing the number of filtered motifs for HcDNV and background variants separated into those that are found only with the reference allele version of the CRS and not the alternate allele version (Disrupted), and motifs that are found in both the reference and alternate forms of the CRS (Undisrupted). Fisher’s exact test p-value: 4.9 × 10−30, Odds Ratio: 2.68.