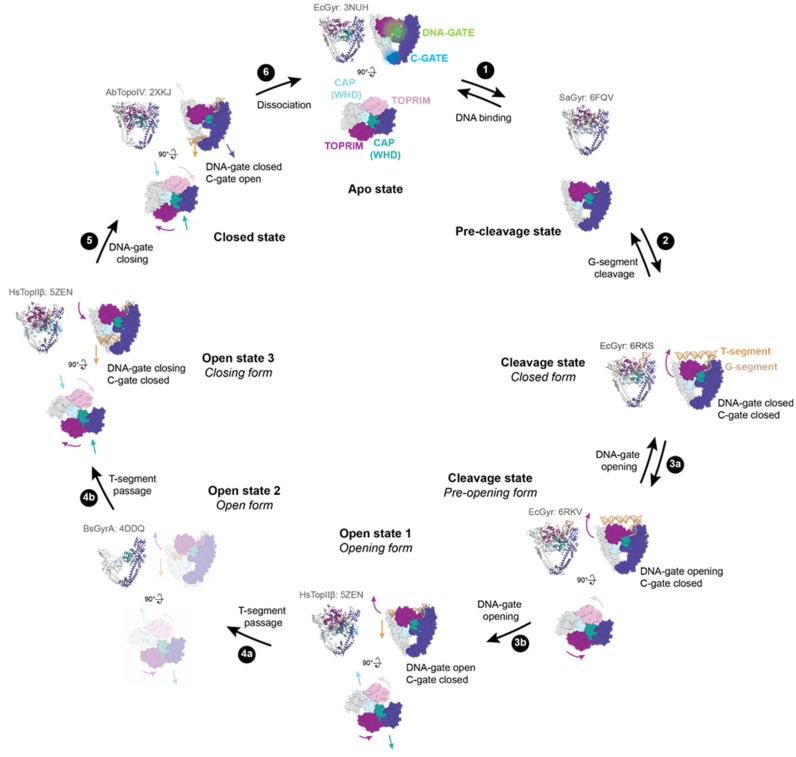
Figure 2.
DNA-gate opening mechanism of type IIA enzymes. All the shown intermediates were structurally determined by X-ray crystallography or cryo-electron microscopy (cryo-EM). PDB accession codes are indicated in light grey. Structure representations restricted to the DNA-binding/cleavage domain were generated from the E. coli DNA gyrase structure without DNA and the insertion domain for clarity. TOPRIM domains are coloured dark magenta and plum. CAP (WHD) domains are coloured light blue and dark cyan. The other domains are coloured in dark slate blue and grey. The G-segment is shown in tan, and the T-segment (modelled) is in sandy brown. (1) G-segment binds to the DNA binding-groove formed by the TOPRIM-CAP(WHD)-Tower domains. (2) DNA binding triggers a rotation of the TOPRIM domains, resulting in a repositioning of the acidic motif with metal ions close to the catalytic tyrosine residues (CAP/WHD) and cleavage of the G-segment. (3) DNA-gate opening involving sliding and swivelling motions (TOPRIM/WHD (CAP)) of the two halves of the protein against each other. 3a and 3b show the transition to the closed form to the pre-opening form of the DNA-gate. The dimer remains together thanks to interactions in the C-gate. (4) From these movements, a funnel-shaped channel is revealed between the two halves of the DNA-gate allowing T-segment passage to change DNA topology. 4a shows the T-segment passage through the upper part of the DNA-gate. The structure of GyrA in an opened conformation suggests that a wider opening of the DNA gate happens to complete strand passage as illustrated in step 4b. (5) After the T-segment passage through cleaved G-segment, the DNA-gate is closed and the G-segment is re-ligated. Once the DNA-gate is closed, the C-gate opens to release the T-segment. (6) After a complete reaction cycle, the enzyme dissociates from the DNA and is reset for another cycle.