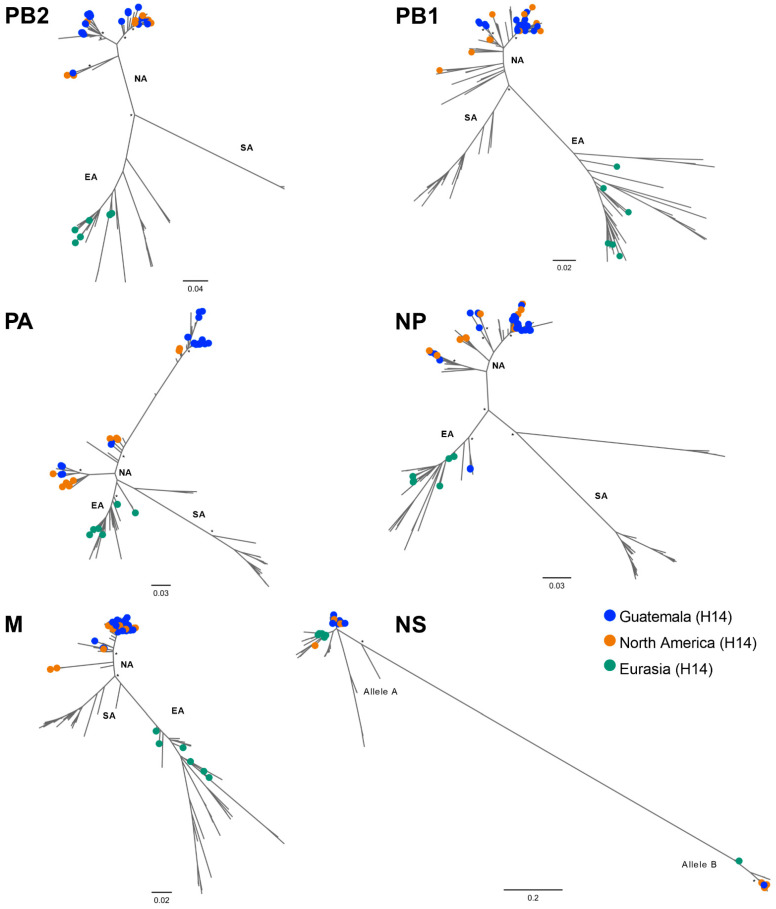
Figure 5.
Internal genes (PB2, PB1, PA, NP, M, and NS) phylogenetic inference for all H14 virus coding sequences available worldwide. Maximum likelihood phylogenetic inference using the best-fit model. Identical H14 viral sequences were removed. Geographic region of H14 viruses is indicated by colored circles. Scale bar on the bottom-left indicates the number of nucleotide substitutions per site. The different lineages from North America (NA), Eurasia (EA), and South America (SA) are shown. Scale bar on the bottom-left indicates the number of nucleotide substitutions per site. Bootstrap values > 70% for key nodes are indicated by asterisks (*) for key nodes.