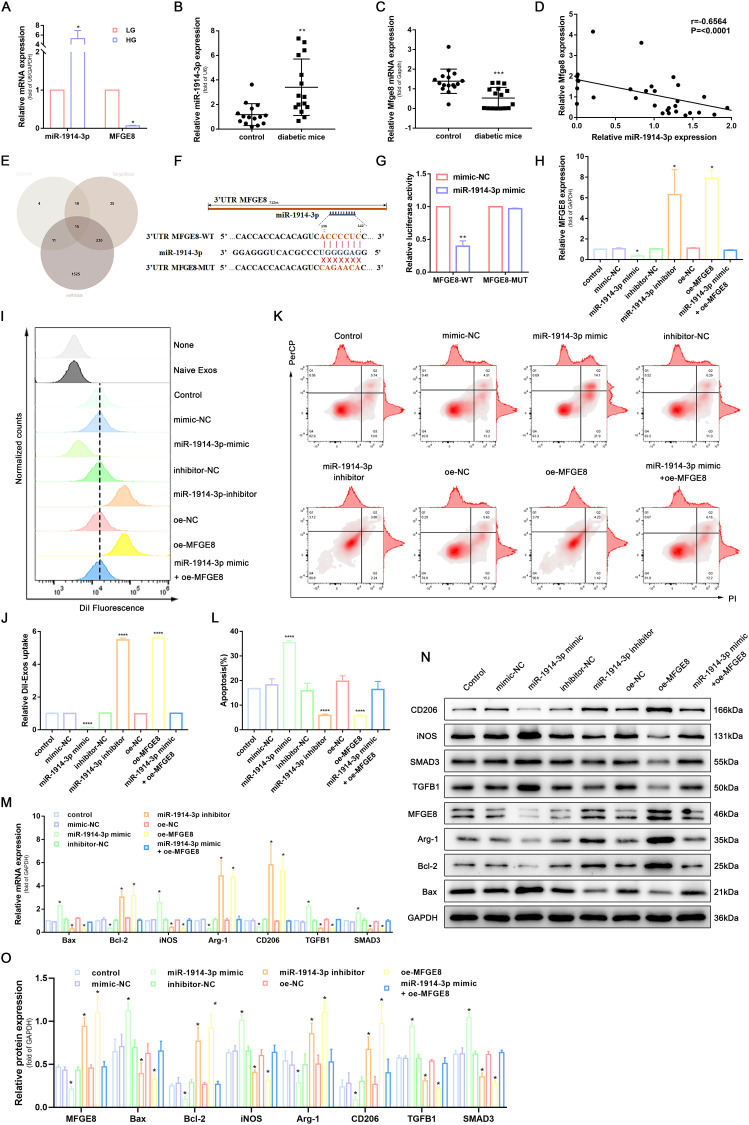
Figure 2.
MiR-1914-3p targeted MFGE8 to affect macrophage phagocytosis, apoptosis, and polarization via the TGFB1/SMAD3 signaling pathway. (A) miR-1914-3p expression and mRNA expression of MFGE8 in THP-1-derived macrophages under a normal medium or high glucose medium, determined by qRT-PCR. (B and C) miR-1914-3p expression and mRNA expression of MFGE8 in wound tissue from normal mice or diabetic mice, determined by qRT-PCR. (D) The correlation between miR-1914-3p and MFGE8 expression, measured in 31 mice skin by Spearman correlation analysis. (E) Predicted target miRNAs of MFGE8 by three online databases (ENCORI, TargetScan and miRWalk). (F) Predicted binding sites between miR-1914-3p and MFGE8 by bioinformatics database. (G) Luciferase activity of MFGE8-WT and MFGE8-MUT detected by dual-luciferase reporter gene assay. (H) mRNA expression of MFGE8, determined by qRT-PCR. (I and J) Uptake of DiI-labelled KCs-Exo by PMA-treated THP-1 cells, measured by flow cytometry analysis. (K and L) Apoptosis in PMA-treated THP-1 cells, measured by flow cytometry analysis. (M) mRNA expression of apoptosis-related factors (Bax and Bcl-2), macrophage polarization-related surface markers (iNOS, Arg-1 and CD206), TGFB1 and SMAD3, determined by qRT-PCR. (N and O) Protein expression of apoptosis-related factors (Bax and Bcl-2), macrophage polarization-related surface markers (iNOS, Arg-1 and CD206), MFGE8, TGFB1 and SMAD3, normalized to GAPDH, determined by Western blot analysis. *p < 0.05, **p < 0.01, ****p < 0.0001 versus the control group. Measurement data were expressed as mean ± SD. Comparison between two groups was conducted using independent sample t test. One-way ANOVA was used for data comparison among multiple groups, followed by Tukey’s post hoc test. The experiment was repeated independently three times.