Figure 1. Comprehensive genetic profiling and validation of somatic variants in 283 MCD patients.
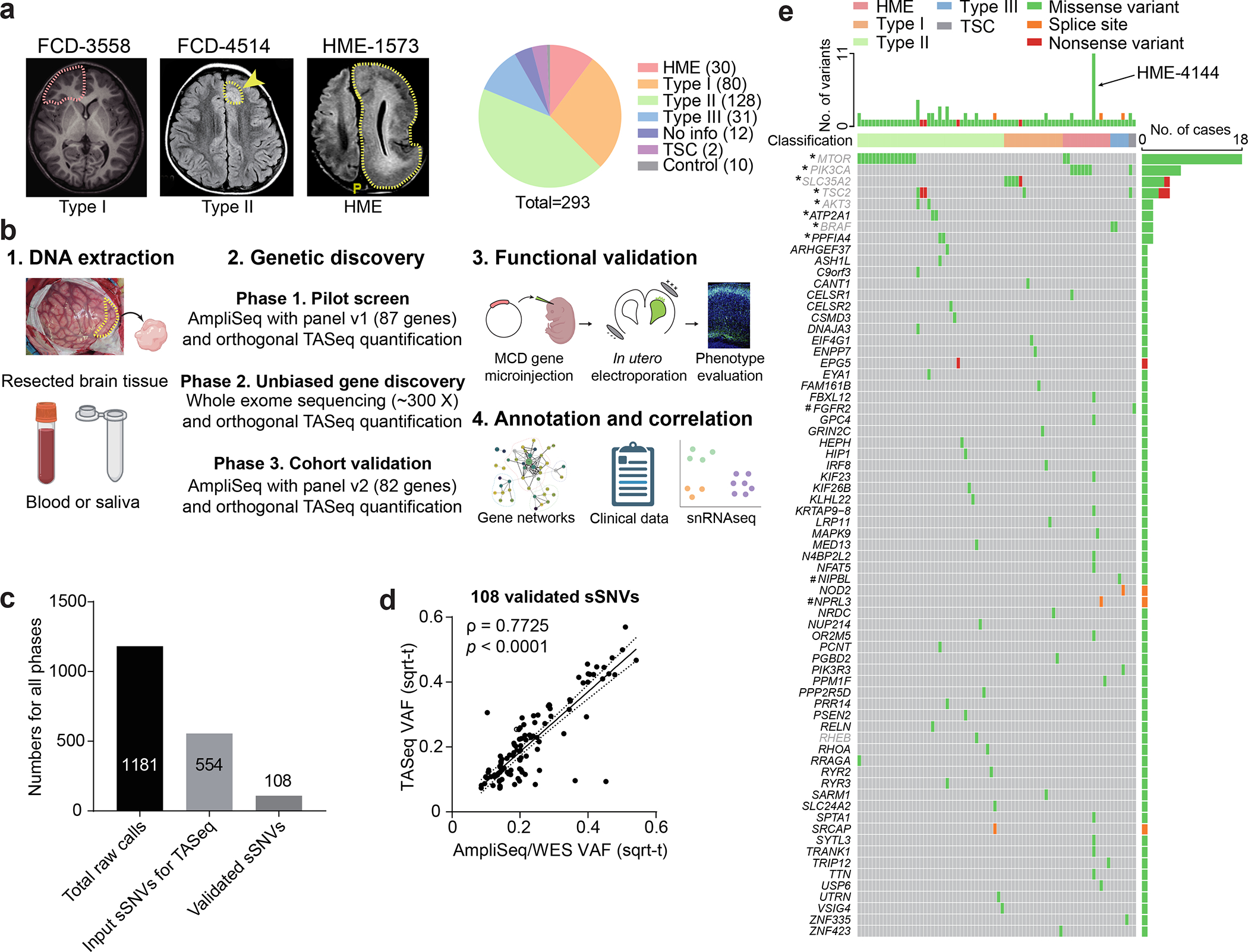
(a) Representative MRI image of FCD-3558 (FCD type I), FCD-4514 (FCD type II), HME-1573, and composition MCD cohort. Yellow arrow and dash: affected brain regions. FCD-3558, MRI was non-lesional, the epileptic focus was mapped to the right frontal lobe (red dashed line) and resected. (b) Three-phase comprehensive genetic MCD profiling workflow, followed by quantification/validation of each variant with target amplicon sequencing (TASeq). Phase 1] 1000 × pilot screening of DNA with an 87-gene mTOR-related panel. Phase 2] 300 × whole-exome sequencing (WES) with best-practice somatic variant discovery for novel candidate genes. Phase 3] Cohort-level validation with an updated, high-confidence TASeq gene set arising from Phases 1 and 2. A subset of somatic mutations was functionally validated in mice. Annotation and correlation with gene networks, clinical data, and single-single-nucleus RNAseq. (c) 1181 sSNV calls were detected from all three phases, yielding 108 validated sSNVs. (d) Correlation between square-root transformed (sqrt-t) AmpliSeq/WES variant allele fraction (VAF) and TASeq VAF. Solid line: Simple linear regression (goodness of fit). Dashed lines: 95% confidence band. Spearman correlation coefficient ρ and corresponding two-tailed t-test p-value. (e) Oncoplot of 69 genes from 108 validated sSNVs. Top: most patients had one gene mutated, a few patients had more than one gene mutated, and patient HME-4144 had 11 different validated genes mutated. Grey: genes recurrently mutated in previous HME/FCD cohorts. *: genes recurrently mutated in our cohort. #: genes non-recurrently mutated but complementary to other cohorts of epilepsy-associated developmental lesions.
