Extended Data Fig. 2. The locations of the selected MCD variants.
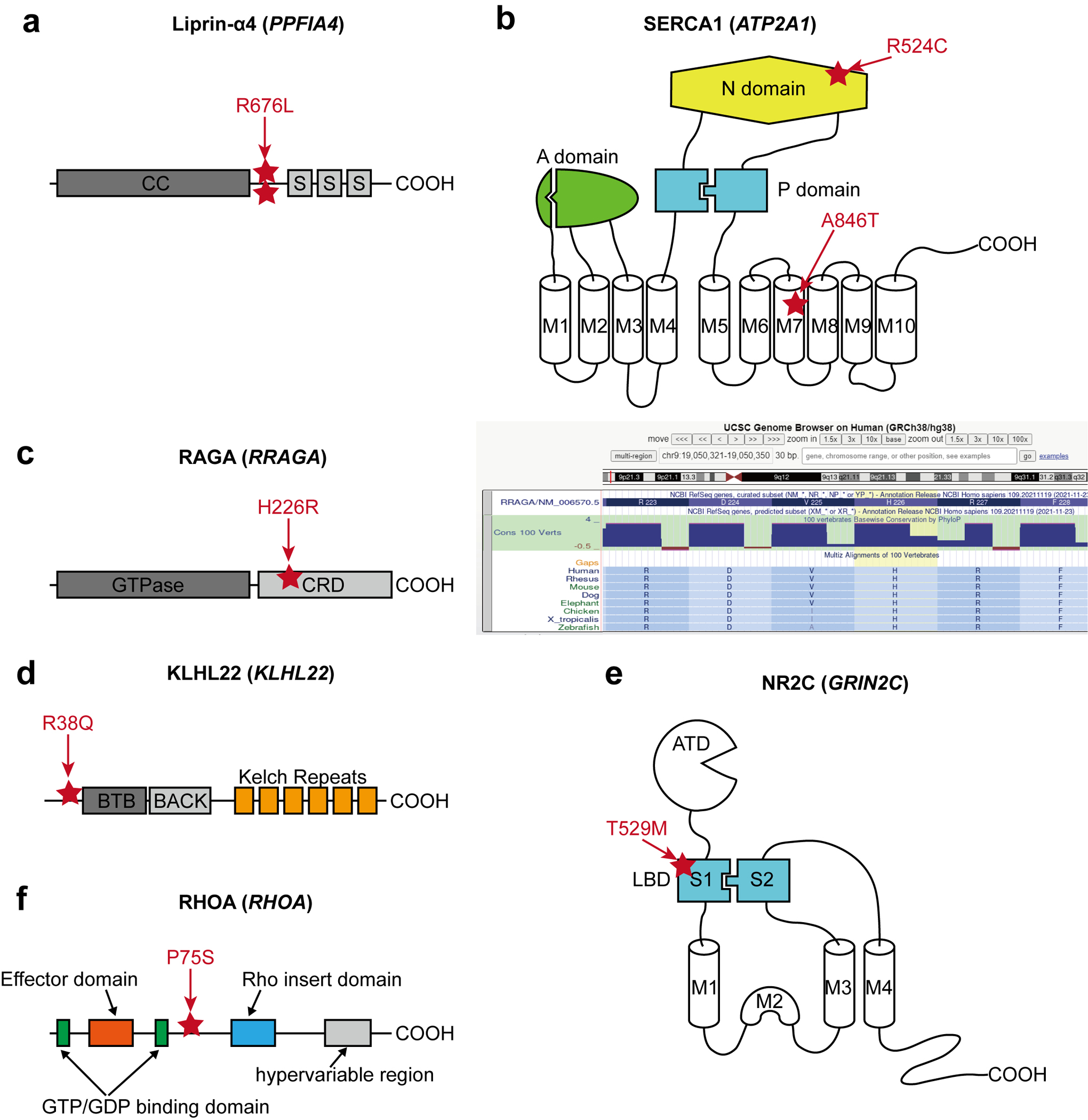
(a) The location of two recurrent SNV calls is at the same position between the coiled-coil domain (CC) and the first SAM domain (S) of Liprin-α4. (b) Two different variants in SERCA1. p.R524C mutation is at the nucleotide ATP-binding (N) domain, whereas the pA846T variant is in the 7th transmembrane (M7) domain. A: Actuator domain, P: Phosphorylation domain, M: Transmembrane domain. (c) Left: The location of p.H226R variant in RAGA protein. GTPase: GTPase domain, CRD: C-terminal roadblock domain. Right: UCSC genome browser screenshot describing that p.H226 is a conserved site across all vertebrates. (d) The location of the p.R38Q variant in the N-terminal region before the BTB (Broad-Complex, Tramtrack, and Bric-à-brac) domain of KLHL22. (e) A variant in the S1 domain of NR2C. S1 and S2 together make the ligand-binding domain (LBD), the target of glutamate. ATD: Amino-terminal domain. (f) RHOA p.P75S variant in the interdomain space between the second GTP/GDP binding domain and Rho insert domain.
