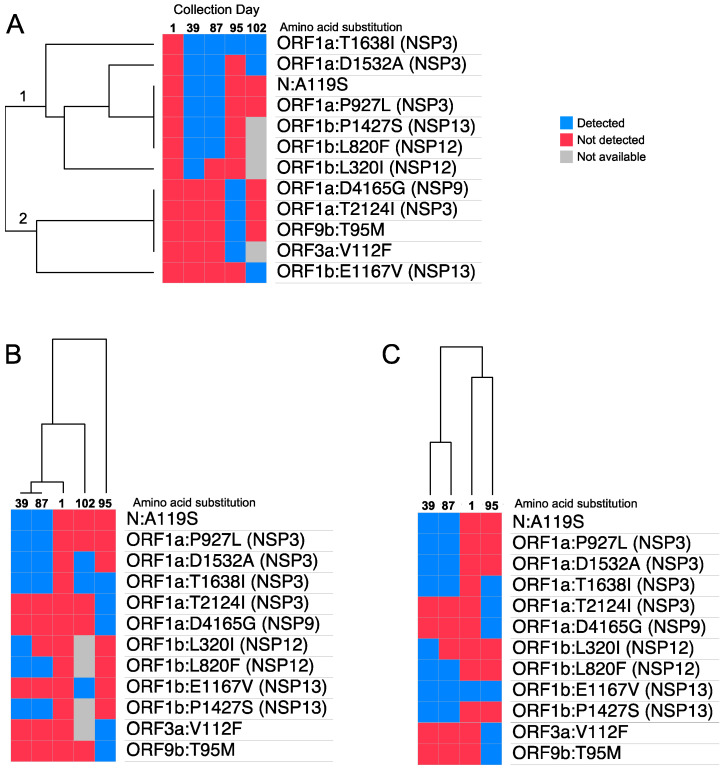
Figure 2.
SARS-CoV-2 hierarchical clustering gene expression map. Euclidean distance was used as the distance metric in the hierarchical clustering algorithm. The colored maps show amino acid substitutions with at least one change within days of nasopharyngeal swab collection. Not available means that the region could not be covered during sequencing, so it does not have mutation data. Panel (A). Hierarchical clustering was applied over mutations to cluster mutations with similar temporal patterns, and isolates were sorted by collection date. Panel (B,C). Hierarchical clustering was applied over collection days to cluster isolates with similar mutation profiles, and mutations were sorted by genomic position. In panel (C), day 102 was removed from the clustering since 4 out of 12 mutations corresponded to missing data, which diminishes the clustering confidence.