Fig. 4.
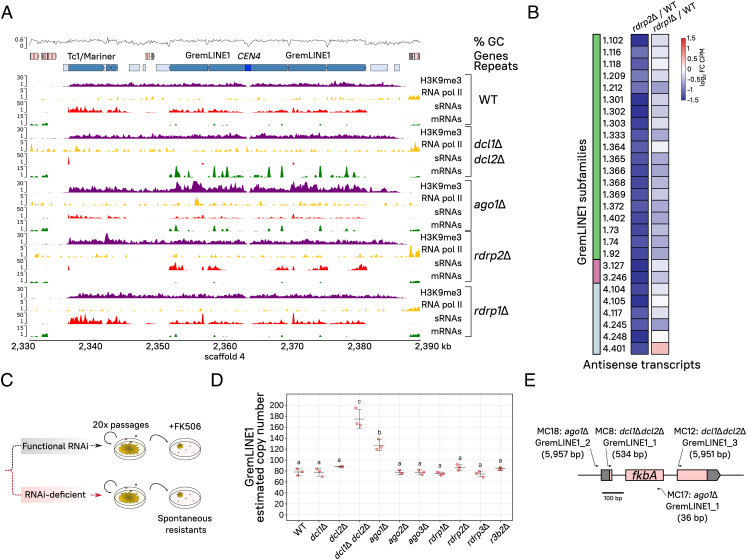
H3K9me defines pericentric boundaries while canonical RNAi inhibits GremLINE1 transposition to noncentromeric regions. (A) Genomic plot of CEN4 and its pericentric regions displaying H3K9me3 (purple) and RNA pol II enrichments (yellow), and sRNA (red) and transcript levels (mRNA, green) in a WT and canonical RNAi-deficient strains. Read coverage is normalized to log2 CPM, and ChIP enrichment is shown as the IP/Input DNA ratio. GC proportion (black line) is computed across the whole sequence. Genes (light red), repeated sequences (structural repeats in light blue and TE in turquoise), as well as the centromeric sequence (dark blue) are shown for reference. (B) Heatmap of significant antisense transcription differences across GremLINE1 copies (FDR ≤ 0.05) in either rdrp2Δ or rdrp1Δ compared with the WT strain (color-coded values shown as log2 fold change of CPM). GremLINE1 subfamilies (color-coded) and identification are shown. (C) Artificial evolution experiment in RNAi mutants. Briefly, the strains were cultured through 20 vegetative growth passages, and analyzed for GremLINE1 copy numbers (D) or FK506 spontaneous resistance (E). (D) GremLINE1 copies in different RNAi mutants after 20 vegetative passages are shown. Copy numbers are estimated as the relative fold (2−ΔCT) between GremLINE1 (multiple copies) and vma1 (single copy) sequence amplification in triplicate qPCR reactions. Significantly distinct values are indicated with different letters and were calculated by one-way ANOVA and Tukey’s tests. (E) GremLINE1 transposition into the fkbA locus was positively selected on FK506-containing medium. Each insertion was named after their genetic background, the TE inserted, and a systematic number; insertion length (in bp) is also shown after the name. Insertion sites are indicated by arrows, whose direction follows the element orientation with respect to fkbA, and whose width reflects the scaled length of the target site duplication marking the insertion site.