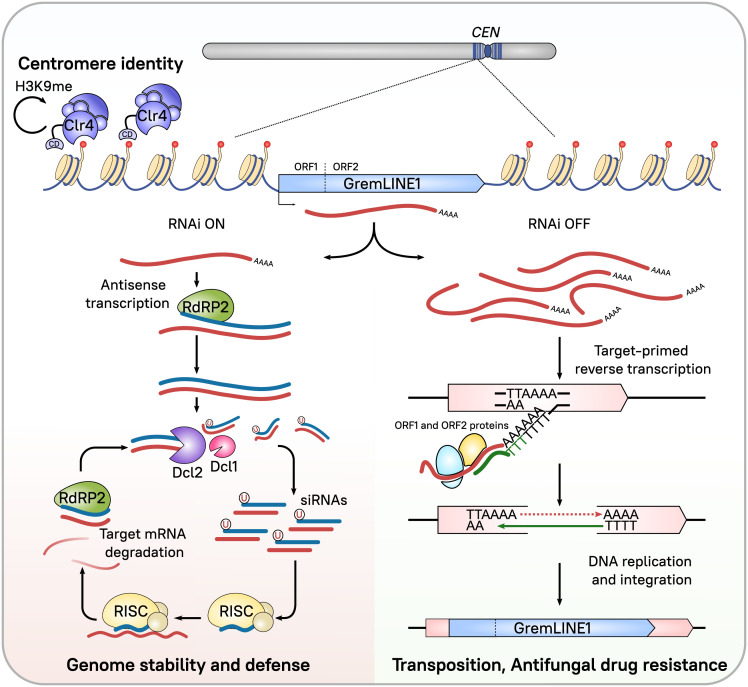
Fig. 5.
Current model for centromeric GremLINE1 dual regulation by H3K9me-based heterochromatin and RNAi. GremLINE1 copies are retrotransposons exclusively located in the pericentric regions of Mucoromycota species, forming long H3K9me heterochromatic islands that define centromere identity. Clr4 methyltransferase catalyzes H3K9me, and its chromodomain allows for H3K9me reading activity that could be involved in maintaining and spreading the mark in an RNAi-independent process. GremLINE1 elements display two ORFs encoding the enzymes necessary for their transposition, and their expression is usually posttranscriptionally repressed by RNAi in a process that involves Rdrp2 antisense transcription, Dcl2 nuclease activity that can be partially rescued by Dcl1, and RISC-targeted degradation of GremLINE1 transcripts. In addition, Rdrp2 activity could reamplify the silencing signal by generating dsRNA from these degraded transcript fragments. This PTGS mechanism ensures genome stability by preventing GremLINE1 transposition and consequently, transposition is increased in RNAi-deficient strains. Lack of RNAi leads to increased GremLINE1 expression and consequently, to target-primed reverse transcription integration events in noncentromeric regions. These transposition events have been shown to affect drug targets, promoting drug resistance.