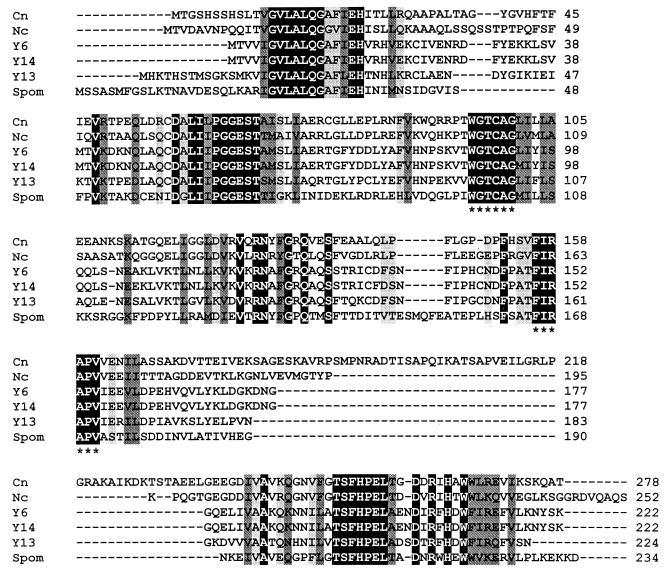
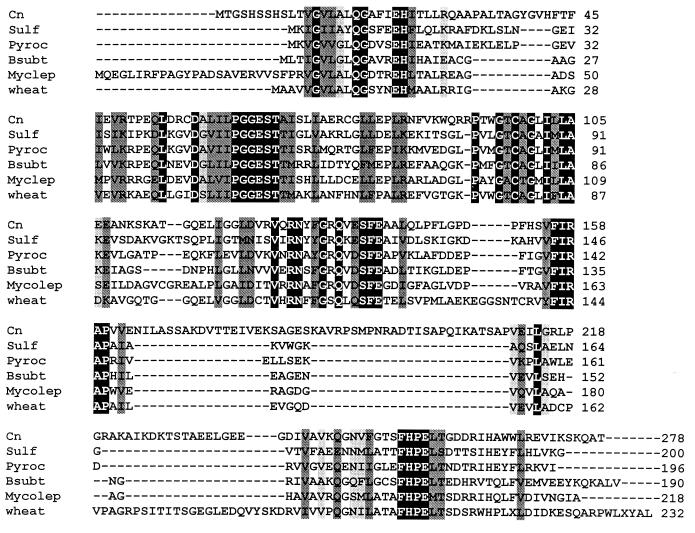
FIG. 4.
Comparison of the predicted amino acid sequences of PDX2 and homologues. (A) Comparison of all fungal PDX2 homologues. Species include C. nicotianae (Cn), Neurospora crassa (Nc), S. cerevisiae homologue from chromosome 6 (Y6), S. cerevisiae homologue from chromosome 14 (Y14), S. cerevisiae homologue from chromosome 13 (Y13), and S. pombe (Spom). The protein sequences from which the degenerate primers were derived are indicated by asterisks. (B) Comparison of the C. nicotianae (Cn) PDX2 protein sequence with representative sequences from other major taxa, including the archaebacteria Sulfolobus solfataricus (Sulf) and Pyrococcus horikoshii (Pyroc), the eubacteria Bacillus subtilis (Bsubt) and Mycobacterium leprae (Myclep), and the plant wheat (Triticum aestivum). The black-boxed residues with white letters indicate identical residues in all proteins, while the darker and lighter shading indicates, respectively, conserved substitutions and semiconserved substitutions.