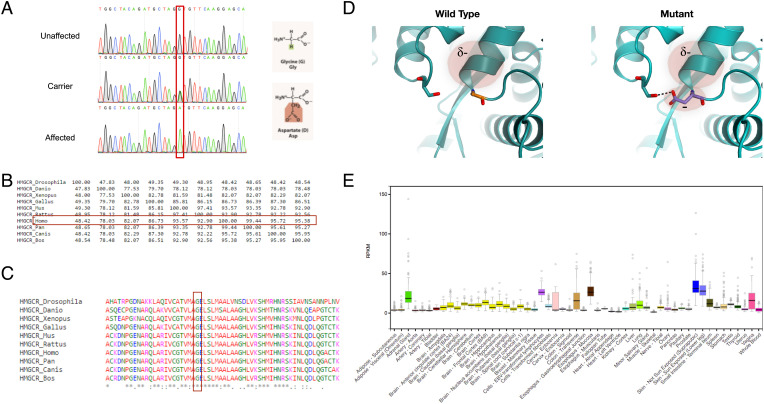
Fig. 2.
The HMGCR mutation. (A) Sanger sequencing of an HMGCR amplicon of an unaffected individual (V:6), an obligatory carrier (IV:1) and an affected individual (V:13). The mutation causes a glycine to aspartate nonconservative substitution at position 822. (B) Precent identity matrix of HMGCR produced using Clustal-Omega, showing high homology between the human HMGCR and orthologs. (C) Multiple alignment of human HMGCR and orthologs produced using Clustal-Omega, showing that the substituted amino acid is highly conserved throughout evolution. (D) Structural model of HMG CoA-reductase protein in the WT and mutated form, based on 1DQ8. The substitution presumably forms a new H-bond with a distal residue, compromises the helix dipole, and causes electrostatic repulsion. (E) Expression of HMGCR across various tissues, obtained using the GTEx database.