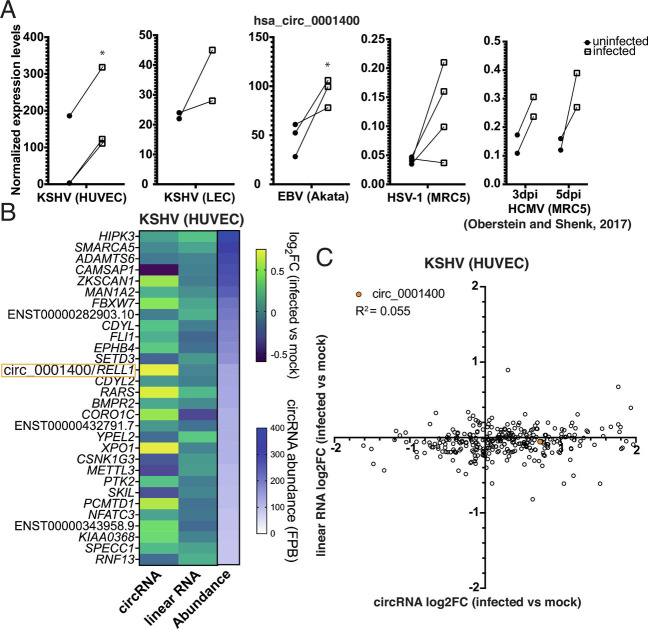
Fig. 1.
Herpesviruses induce circ_001400 expression. (A) hsa_circ_0001400 transcript levels are shown in KSHV-infected HUVECs at 3 d post-infection (dpi) (microarray), LECs at 3 dpi (RNA-Seq), EBV-positive Burkitt’s lymphoma line Akata (microarray), HSV-1-infected fibroblast at 12 h post-infection (RNA-Seq), and HCMV-infected fibroblast at 3 and 5 dpi (RNA-Seq) (16). n = 2 to 4. Significances were computed when n is 2 or more with limma. *P-value < 0.05. (B) Fold-changes of top 30 most abundant human circRNAs in KSHV-infected HUVECs. Transcript levels of circRNAs and corresponding linear messenger RNAs (mRNAs) were quantitated from total RNA-Seq with circExplorer3 (18) as FPBs (fragments per billion mapped base). Fold-changes of circRNAs and linear RNAs (infected vs. mock) are calculated (n = 3), and mean values are shown. Genes are sorted by circRNA FPBs (Abundance), and host genes of both circRNA and linear RNAs are shown on the left. FPBs and ratios are available in Dataset S1. (C) Log2 fold-changes of all detected human circRNAs/counterpart linear mRNAs (KSHV-infected vs. mock HUVECs) are shown as a scatter plot. Data are same as in Fig. 1B and Dataset S1. Pearson correlation value was calculated. n = 3 and mean values are shown.