Fig. 3.
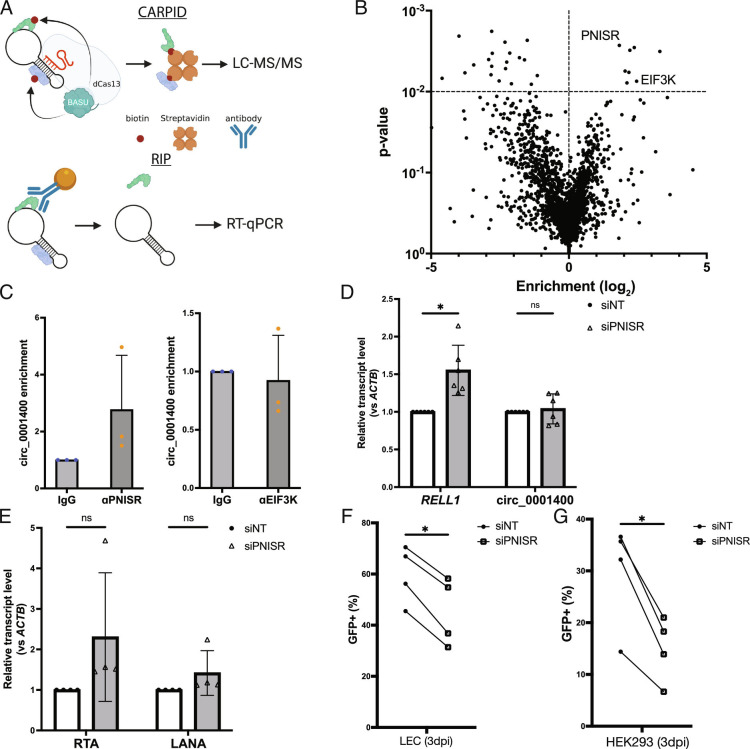
Protein interactome of circ_0001400. (A) Diagram of CARPID and RIP method. CARPID utilizes dead Cas13 conjugated with BASU biotin transferase to biotinylate proteins that are in the proximity of circ_001400 to enrich and determine their identities by liquid chromatography/tandem mass spectrometry. Identified proteins are listed in Dataset S2. Candidate proteins are immunoprecipitated by specific antibodies (RIP), and circ_001400 is confirmed to be co-precipitated by RT-qPCR. (B) A volcano plot of proteins that were identified by mass spectrometry (n = 3). Enrichment and significances were calculated by RankProd package. Pre-gRNA (empty) vector and gCirc1400_nc (sequence-randomized) vector were used as negative controls. (C) RIP results of candidate proteins. circ_0001400 was ectopically expressed in 293T cells for 24 h, and RIPs were performed against PNISR and EIF3K. circ_001400 transcript levels after RIP are normalized to the isotype IgG antibody condition. n = 3 and significances were calculated with paired-t tests. *P-value < 0.05. (D and E) PNISR knockdown and quantitation of RELL1 transcripts and viral transcripts. LECs were transfected with siRNAs targeting PNISR followed by KSHV infection at MOI of 1.0 i.u for 3 d. Linear and circular transcripts from RELL1 gene locus (RELL1 and circ_000400) (D) and viral transcripts (E) were quantified. N = 4 to 5. (F) Infection rates of PNISR-depleted LEC cells. LECs from (D and E) were quantified for the green fluorescent protein (GFP) by flow cytometry. N = 4. (G) Virion production of PNISR-depleted LEC cells. Conditioned media was collected from LECs from (F), and HEK-293 cells were incubated with the conditioned media for 3 d. Infected HEK-293 cells were counted by GFP signals with flow cytometry. N = 4 and significances were calculated with paired-t tests. *P-value < 0.05.