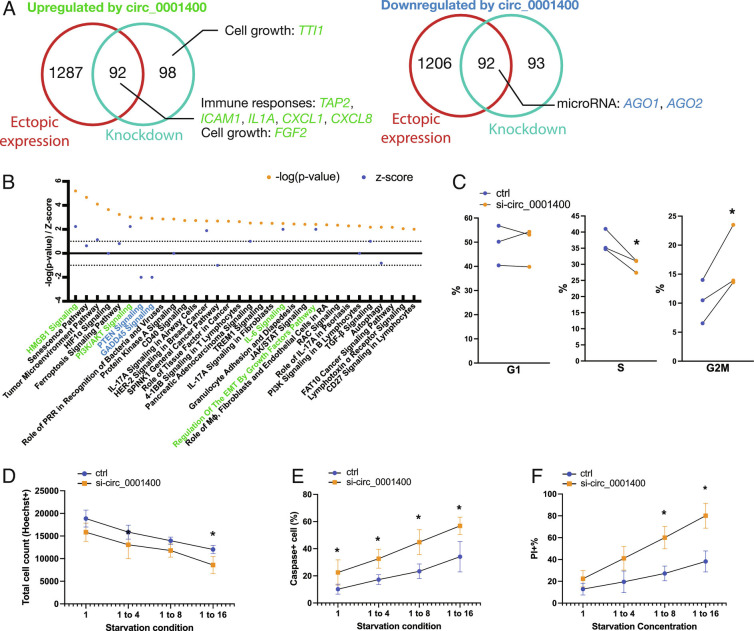
Fig. 5.
circ_001400 promotes cell growth and induces immune reaction genes. (A) Venn diagrams are showing consistently differentially regulated genes (DEGs) by circ_0001400 in HUVECs. Cells were treated with siRNA targeted against circ_0001400 (n = 3) or infected with circ_0001400-expressing lentivirus (n = 3) followed by KSHV infection. DEGs are defined as having adjusted P-value <0.05 as calculated by voom (limma package). On 3 d post-infection, the total RNA was extracted for RNA-Seq. Select up-regulated (left Venn diagram) or down-regulated (right Venn diagram) genes by circ_0001400 are shown in green and blue, respectively. The list of DEGs is available in Dataset S4. (B) Pathway enrichment analysis of DEGs (A) by Ingenuity Pathway Analysis (Qiagen). Significance of pathway enrichment is shown as P-values. Z-scores describe the predicted activation or inhibition of pathways. Pathways are color-coded when |Z| > 1.96. (C) Cell cycle analysis of circ_0001400-depleted KSHV-infected cells. Infected cells were incubated with EdU at 3 d post-infection for 4 h, and cell cycles were quantitated by flow cytometry (n = 3). (D–F) Total, apoptotic, and dead cell quantitation of circ_0001400-depleted KSHV-infected cells. HUVECs were siRNA-treated for circRNA depletion and infected with KSHV for three days. The next day, the media was replaced with diluted media to the ratio as described. At 3 d post-infection, total cells were counted by Hoechst staining. Caspase3/7 substrates that emit signal upon cleavage were added to measure apoptosis rates. Dead cells were stained with propidium iodide. Cells were quantitated by high-content imaging (n = 3). Significances were calculated with paired-t tests. *P-value < 0.05.