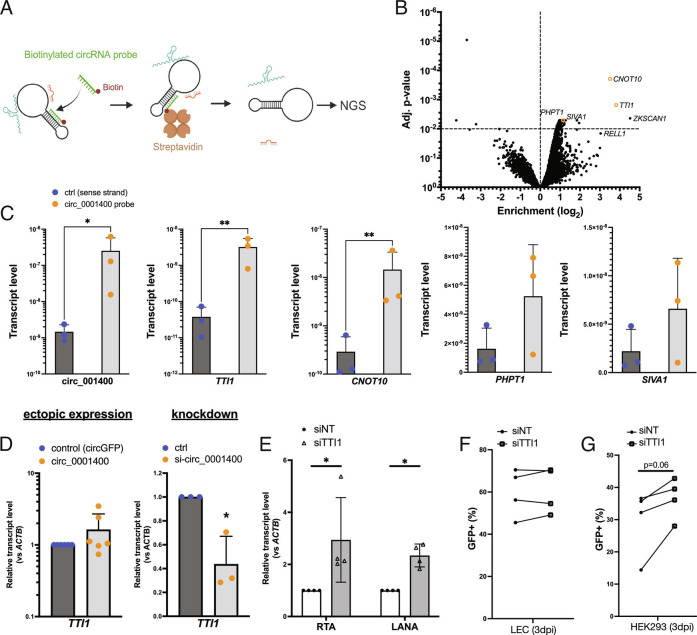
Fig. 7.
circ_001400 interacts with mTOR-component TTI1 and sustains its transcript level. (A) Diagram of the RNA-pulldown assay is shown. Total RNA was incubated with biotinylated DNA oligos that is specific to circ_0001400, and circRNA-RNA complexes are purified using streptavidin beads. Next-generation sequencing was performed to determine their identities. (B) A volcano plot of transcripts identified by the circ_0001400 RNA-pulldown assay in 293T cells. Mapped reads were median scaled, and enrichments were calculated by voom (limma package). ZKSCAN1 and RELL1 are partially included in the circ_0001400 expression plasmid vectors and thus serve as positive control of pulldown. Transcripts that were confirmed in Fig. 6C are depicted as orange rectangles. n = 3. (C) RT-qPCR of enriched transcripts after RNA-pulldown. circ_0001400 was ectopically expressed in 293T cells for 24 h, and RNA-pulldowns were performed (n = 3). Enrichment of circ_0001400 and candidate mRNAs (listed in Dataset S5) are shown. (D) TTI1 transcript levels after manipulation of circ_0001400 transcript levels. circ_0001400 was ectopically expressed (n = 6) or depleted by siRNAs (n = 3) in 293T cells for 48 h. Total RNAs were extracted followed by RT-qPCR. (E) RNAi-mediated knockdown of TTI1. LECs were transfected with siRNAs targeting TTI1 followed by KSHV infection at MOI of 1.0 i.u. RNA was extracted 3 d post–infection, and transcript levels of viral transcripts were measured. N = 4. (F) Infection rates of TTI1-depleted LEC cells. LECs from (E) were quantified for GFP by flow cytometry. N = 4. (G) Virion production of TTI1-depleted LEC cells. Conditioned media was collected from LECs from (F), and HEK-293 cells were incubated with the conditioned media for 3 d. Infected HEK-293 cells were counted by GFP signals with flow cytometry. N = 4 and significances were calculated with paired-t tests. *P-value < 0.05.