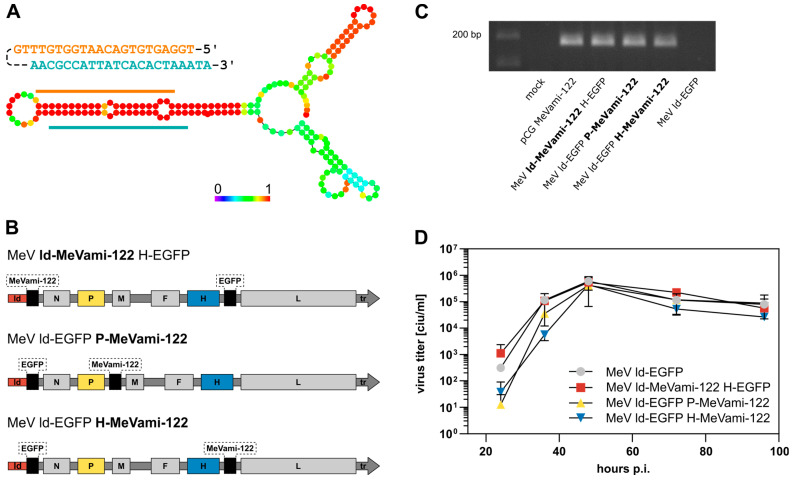
Figure 1.
Design of MicroRNA-encoding MeV. (A) Depiction of MeVami-122 expression cassette. The pre-miR-122 sequence comprises the miR-122 sequence (orange) and its complementary target site sequence (blue). The precursor sequence was elongated by 50 nucleotides 5′, and 49 nucleotides 3′ of pri-miR-122, ensuring both the recognition by the microprocessor and the polyhexameric length of the viral genome upon integration of the cassette. Below: Secondary structure of the MeVami-122 cassette, as predicted by the “RNAfold webserver” of the University of Vienna [29]. Colors indicate base pairing probabilities, wherein red indicates a base pairing probability of 100%. (B) Schematic depiction of the three MeV genomes harboring the MeVami-122 cassette. EGFP and MeVami-122 transgenes were inserted at the indicated positions. (C) Transgene expression of MeVami-122-encoding MeV. Vero cells were either transfected with pCG MeVami-122 or infected with one of the MeVami-122 encoding viruses. Samples from RT-PCR performed with pri-miR-122 specific primers were run on an agarose gel. (D) Virus growth curves. Vero cells were infected with the indicated recombinant MeV variant (MOI 0.03) and titers of progeny virus were determined at the indicated time points. The mean of n = 2 experiments is plotted. Error bars show standard deviation.