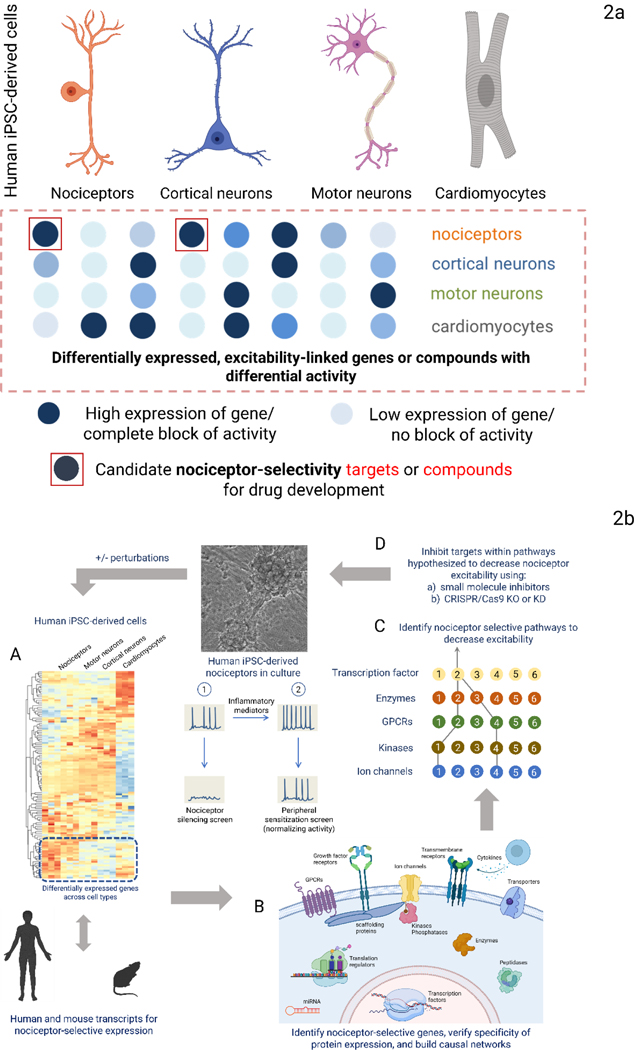
Figure 2:
Fig. 2a: Two complementary approaches to identify novel analgesic targets. Human iPSC-derived nociceptors can be used to identify novel analgesic targets. In one approach, differential gene expression in nociceptors is identified. In the second, an annotated library of compounds, whose primary targets of action are known, are screened for decreases in activity on multiple cell types. The annotated targets of compounds that selectively only inhibit nociceptor activity are potential pain targets. Fig. 2b: Identifying and validating candidate nociceptor-selective targets and pathways. Genes selectively expressed in iPSC-derived nociceptors can be identified (A). A similar comparison of primary mouse and human tissues will yield sets of genes selectively expressed in nociceptors for each species. These can be assembled into signaling pathways (B) predicted to affect nociceptor excitability selectively (C) and represent potential translatable target sets. Nociceptor-selective targets can be validated using small molecules that selectively modulate the function of targets or through CRISPR/Cas9 editing (D). The process can be repeated in the presence of inflammatory mediators, chemotherapeutic agents, or after axon injury to discover selective targets for each condition. Reporters of intracellular signaling can be used to garner information on pathways activated by disease perturbations.