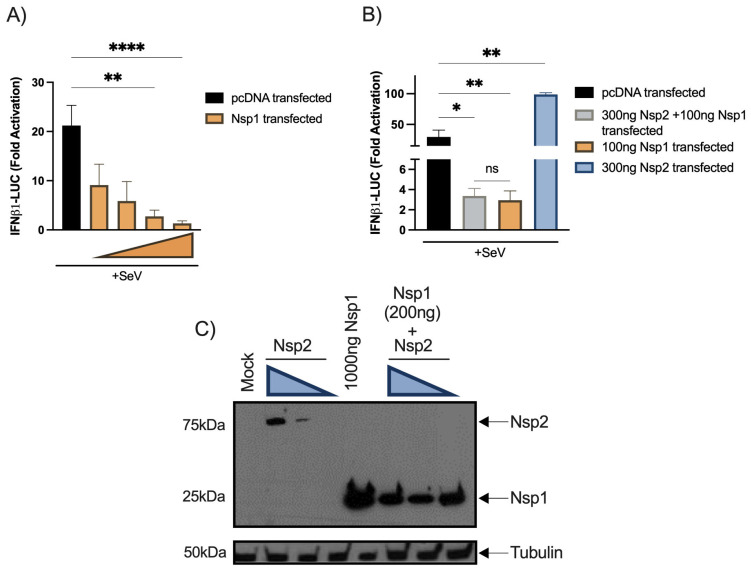
Figure 5.
Effect of SARS-CoV-2-Nsp1 and Nsp1-Nsp2 cotransfection on SeV-induced IFNβ1 promoter activation. HEK293T cells were seeded, transfected, and simulated according to the procedure described above and the luciferase activity was measured then standardized as described for other reporter assays. Doses of 300 ng, 100 ng, 30 ng, and 3 ng per well of Nsp1 vectors for Nsp1 alone experimentation (A). HEK293T cells were seeded, transfected, and simulated according to the procedure described above and the luciferase activity was measured then standardized as described for other reporter assays. Doses of 300 ng of Nsp2 aand 100 ng of Nsp1 per well were used (B). All experiments were performed twice in triplicate and the compilation of the data is shown. Results are expressed as fold activation relative to the pcDNA mock-infected control (mean ± SD, n = 6 replicates/condition). The Nsp1-Nsp2 cotransfection and Nsp1 transfection conditions were compared to the mock-transfected control using nonparametric one-way ANOVA with Dunn’s correction and Nsp2 conditions were compared using nonparametric one-way ANOVA. The Nsp1-Nsp2 cotransfection and Nsp1 transfection conditions were compared using nonparametric t-test. * p < 0.05, ** p < 0.01, **** p < 0.0001, ns: not significant. Protein expression of Nsp1 and Nsp2 in individual transfection and cotransfection (C). HEK293T were seeded in 6-well plate. Twenty-four hours after, cells were transfected with control vector or Nsp1 expression vector or Nsp2 expression vector. Doses of 800 ng, 500 ng, and 100 ng of Nsp2 vector were used. Forty-eight hours post-transfection, cells were lysed in SDS PAGE 2X buffer and detected by Western Blot with an anti-FLAG (viral protein) and rabbit anti-tubulinβ (loading control) on radiological films. The Western Blot was performed two times and one representative experiment is shown.