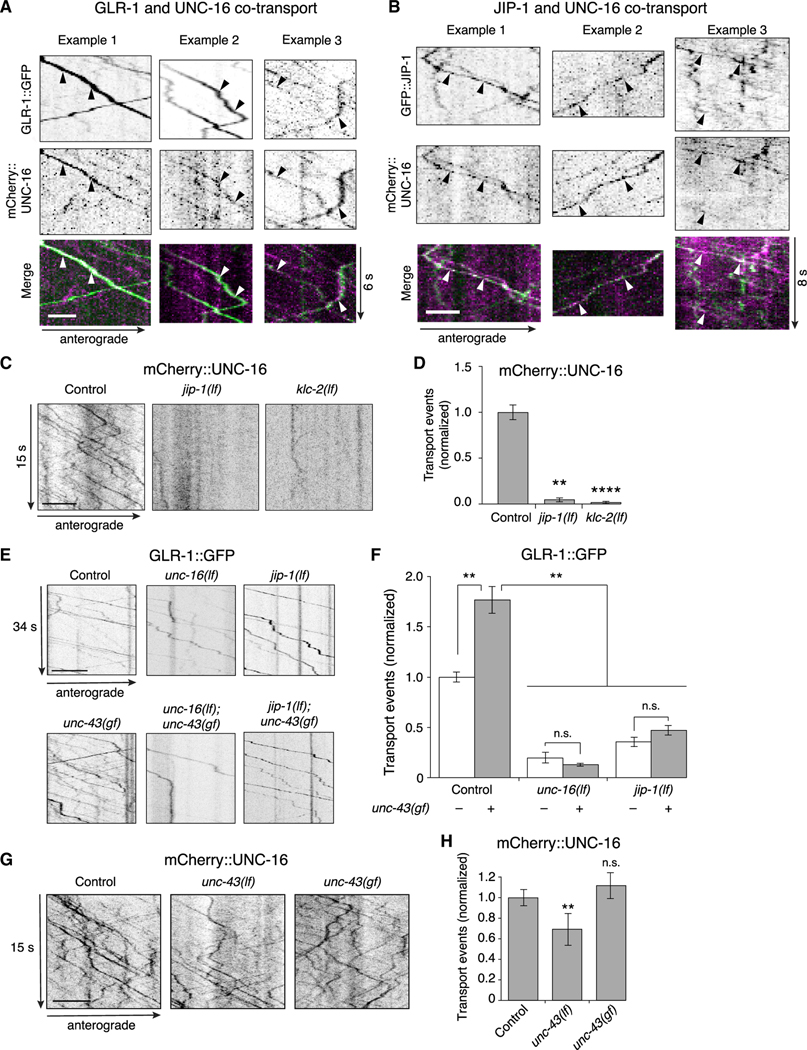
Figure 4. The UNC-16 and JIP-1 scaffold proteins are co-transported and are required for CaMKII-dependent AMPAR transport.
(A and B) Kymographs showing co-transport events (arrowheads) of mCherry::UNC-16 with either GLR-1::GFP (A), or GFP::JIP-1 (B) along the AVA processes. Scale bars, 2 μm.
(C) Kymographs of mCherry::UNC-16 transport in the AVA processes of transgenic control worms, and jip-1(lf) and klc-2(km11) lf mutants. All strains carried the unc-16(e109) mutation. Scale bar, 5 μm.
(D) Total mCherry::UNC-16 transport events in control (n = 9), jip-1(lf) (n = 11), and klc-2(lf) (n = 10). Significantly different from control, **p < 0.01 and ****p < 0.0001 (Kruskal-Wallis and Dunn’s multiple testing).
(E) Kymographs of GLR-1::GFP transport in the AVA processes of transgenic control worms, and various single and double mutants. Scale bars, 5 μm.
(F) Total GLR-1::GFP transport events in control worms (n = 9), unc-16(lf) (n = 9), jip-1(lf) (n = 14), and unc-43(n498sd) gain-of-function (gf) (n = 12) single mutants, and unc-43(gf); unc-16(lf) (n = 6) and jip-1(lf); unc-43(gf) (n = 6) double mutants (normalized to control without unc-43(gf)). Strains with a wild-type unc-43 allele or unc-43(gf) allele are indicated by white and gray bars, respectively. **p < 0.01; n.s., not significant using ANOVA with Kruskal-Wallis and Dunn’s multiple testing.
(G) Kymographs of mCherry::UNC-16 transport in transgenic control worms, unc-43(n498 n1186) lf, and unc-43(gf) mutants. All strains carried the unc-16(e109) mutation. Scale bars, 5 μm.
(H)Total mCherry::UNC-16 transport events in control (n = 13), unc-43(lf) (n = 9), and unc-43(gf) (n = 4). **Significantly different from control; p < 0.01; n.s., not significantly different from control (Mann-Whitney U test between samples acquired the same day).
Error bars represent SEM. See STAR Methods for a description of statistical analysis. See also Figure S5, Tables S1, and S2.