Abstract
The complete sequencing of bacterial genomes has revealed a large number of drug transporter genes. In Escherichia coli, there are 37 open reading frames (ORFs) assumed to be drug transporter genes on the basis of sequence similarities, although the transport capabilities of most of them have not been established yet. We cloned all 37 putative drug transporter genes in E. coli and investigated their drug resistance phenotypes using an E. coli drug-sensitive mutant as a host. E. coli cells transformed with a plasmid carrying one of 20 ORFs, i.e., fsr, mdfA, yceE, yceL, bcr, emrKY, emrAB, emrD, yidY, yjiO, ydhE, acrAB, cusA (formerly ybdE), yegMNO, acrD, acrEF, yhiUV, emrE, ydgFE, and ybjYZ, exhibited increased resistance to some of the 26 representative antimicrobial agents and chemical compounds tested in this study. Of these 20 ORFs, cusA, yegMNO, ydgFE, yceE, yceL, yidY, and ybjYZ are novel drug resistance genes. The fsr, bcr, yjiO, ydhE, acrD, and yhiUV genes gave broader resistance spectra than previously reported.
Bacterial species that have developed clinical resistance to antimicrobial agents are increasing in numbers, and the mechanisms underlying their resistance are being studied. The active efflux of antibiotics is mediated by a family of transmembrane proteins frequently referred to as drug resistance translocases (3, 31, 39, 40, 53). The first drug-pumping protein to be reported was the plasmid-encoded tetracycline resistance Tet protein in 1980 (24).
Five families of drug extrusion translocases are currently identified based on sequence similarity (3, 31, 42, 53). These are the MF (major facilitator) family, SMR (small multidrug resistance) family, RND (resistance nodulation cell division) family, ABC (ATP-binding cassette) family, and recently identified MATE (multidrug and toxic compound extrusion) family (4). Membrane transporters of the MF family possess 12 to 14 transmembrane domains. For example, Bcr (Escherichia coli) (1), EmrB (E. coli) (19), EmrD (E. coli) (29), MdfA/Cmr (E. coli) (6, 32), NorA (Staphylococcus aureus) (58), QacA (S. aureus) (45), and Bmr (Bacillus subtilis) (30) are members of this family, and these systems mediate drug extrusion with different specificities. Transporters of the SMR family are rather small and usually possess four transmembrane domains. SMR (or QacC) (S. aureus) (11, 18), QacE (Klebsiella aerogenes) (35), and EmrE (E. coli) (57) belong to this family. EmrE seems to be organized as a homooligomer, most likely a trimer (28). Some proteins in this family appear to function as heterooligomers (14, 23). Transporters of the RND family are usually components of tripartite efflux systems facilitating extrusion of substrates directly into the external medium rather than into the periplasm. Besides the inner-membrane RND transporter, such systems contain a membrane fusion protein (MFP) and an outer membrane factor (OMF). AcrA-AcrB-TolC (E. coli) (21) and MexA-MexB-OprM (Pseudomonas aeruginosa) (41) are examples of tripartite efflux systems, where AcrB and MexB are RND transporters, AcrA and MexA are MFPs, and TolC and OprM are OMFs. An electrochemical potential gradient of H+ across cell membranes seems to be the driving force for drug efflux by the MF, SMR, and RND family transporters (9, 59). Transporters of the ABC family utilize ATP as an energy source. LmrA (Lactococcus lactis) (54) and MsrA (S. aureus) (44) are members of this family. Transporters of the MATE family have 12 predicted transmembrane segments, like members of the MF family. However, the MATE family transporters do not exhibit significant sequence similarity to any member of the MF family (4). NorM (Vibrio parahaemolyticus) (26) is a member of this family. NorM seems to be an Na+-driven multidrug efflux pump (25).
The last 5 years have seen impressive progress in the sequencing of the entire genomes of both prokaryotic and eukaryotic free-living organisms. As of June 2001, the complete genome sequences of 46 microbial species were publicly available (TIGR microbial database, http://www.tigr.org/tdb/mdb /mdbcomplete.html). The complete genome sequence of E. coli was determined by Blattner et al. in 1997 (2). Surprisingly, analysis of microbial genomes has revealed a large number of putative drug transporter genes (36–38, 48). In E. coli, in the five families, 37 putative drug transporter genes (19 MF, 3 SMR, 7 RND, 7 ABC, and 1 MATE) were found in the course of sequence annotation. The transport systems of these genes were classified according to the putative membrane topology, protein family, bioenergetics, and substrate specificity (38). However, the transport capability of the majority of them has not been established yet.
To investigate the power of prediction and to investigate the substrate specificity of potential drug transporters, first we cloned all 37 putative drug transporter genes of E. coli with native promoters and proximate genes into multicopy plasmids and then investigated their drug resistance phenotypes using an E. coli mutant lacking the major multidrug efflux system AcrAB as a host. We examined the susceptibility of E. coli cells harboring a multicopy plasmid carrying a putative drug transporter gene to 26 representative antimicrobial agents and chemical compounds well translocated by AcrAB or other major drug transporters. We found that 16 of 33 plasmids constructed conferred drug resistance on E. coli cells. The other 17 of the 33 multicopy plasmids carrying drug transporter open reading frames (ORFs) did not produce increased resistance to any compounds tested in this study. There is a possibility that these ORFs might not be expressed from their own native promoters or that their expression might be repressed by other ORFs that were cloned simultaneously. For this reason, we cloned these ORFs into an expression vector and then investigated their drug resistance phenotypes with induction by IPTG (isopropyl-β-d-thiogalactopyranoside). As a result, we identified several new drug resistance genes by using information on the complete genome sequence, and we showed that the substrate spectra of some previously identified transporters were more extensive than originally thought.
MATERIALS AND METHODS
Bacteria and growth.
E. coli W3104 (56) was used as the donor of chromosomal DNA. E. coli TG1 (50) was used as the cloning host. E. coli KAM3 (26), a derivative of K-12 that lacks a restriction system and AcrAB, was used for drug susceptibility testing and expression confirmation. E. coli cells were grown in 2×YT medium (46), supplemented with ampicillin (100 μg/ml) when necessary, under aerobic conditions at 37°C. Competent cells were prepared by the method of Hanahan (12).
Drug susceptibility test.
The MICs of drugs were determined on YT agar containing various drugs (chloramphenicol, tetracycline, minocycline, erythromycin, nalidixic acid, norfloxacin, enoxacin, kanamycin, vancomycin, fosfomycin, fosmidomycin, bicyclomycin, doxorubicin, novobiocin, rifampin, trimethoprim, acriflavine, crystal violet, ethidium bromide, rhodamine 6G, methylviologen, tetraphenylphosphonium bromide [TPP], carbonyl cyanide m-chlorophenylhydrazone [CCCP], benzalkonium, sodium dodecyl sulfate [SDS], and deoxycholate) at various concentrations, as indicated. These agar plates were made by the twofold agar dilution technique (33). We added 0.1 mM IPTG to agar plates when we examined the susceptibility of E. coli cells harboring pTrc6His plasmids that carry drug transporter ORFs under control of the trc promoter (see Tables 1 and 3). A total of 104 cells on a test agar plate were incubated at 37°C for 24 h, and then growth was evaluated.
TABLE 1.
Plasmids used in this studya
| Plasmid | Relevant characteristic | Origin |
|---|---|---|
| General | ||
| pUC118 | Vector: Apr; multiple cloning site in lacZ | Takara Shuzo Co. |
| pUC119 | Vector: Apr; multiple cloning site in lacZ | Takara Shuzo Co. |
| pTrc99A | Expression vector: Apr; multiple cloning site downstream of trc promoter | Amersham Pharmacia Biotech |
| pTrc6His | pTrc99A-derived expression vector: Apr; C-terminal 6-His tag | This study |
| Plasmids carrying MF-type transporter ORFs | ||
| pUCyajR | 2.0-kb KpnI-HindIII fragment containing yajR (putative transporter) gene cloned into pUC118, Apr | This study |
| pUCfsr | 1.8-kb EcoRI-SphI fragment containing fsr (fosmidomycin transporter) gene cloned into pUC118, Apr | This study |
| pUCmdfA | 1.6-kb KpnI-HindIII fragment containing mdfA (multidrug transporter) gene cloned into pUC118, Apr | This study |
| pUCyceE | 1.4-kb SphI-BamHI fragment containing yceE (putative transporter) gene cloned into pUC119, Apr | This study |
| pUCyceL | 1.7-kb SphI-BamHI fragment containing yceL (putative transporter) gene cloned into pUC119, Apr | This study |
| pUCydeA | 1.4-kb SphI-BamHI fragment containing ydeA (l-arabinose and IPTG transporter) gene cloned into pUC119, Apr | This study |
| pUCydeF | 1.7-kb SphI-BamHI fragment containing ydeF (putative transporter) gene cloned into pUC119, Apr | This study |
| pUCynfM | 2.5-kb SphI-BamHI fragment containing ynfL (putative regulator) and -M (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCydhC | 2.4-kb SphI-BamHI fragment containing ydhB (putative regulator) and -C (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCydiM | 1.7-kb SphI-BamHI fragment containing ydiM (putative transporter) gene cloned into pUC119, Apr | This study |
| pUCyebQ | 2.8-kb BamHI-EcoRI fragment containing kdgR (putative regulator) and yebQ (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCbcr | 2.3-kb AccI-KpnI fragment containing yeiD (putative regulator) and bcr (bicyclomycin transporter) genes cloned into pUC119, Apr | This study |
| pUCemrKY | 7.5-kb SphI-BamHI fragment containing evgS/A (two-component system) and emrK/Y (putative MFP/putative transporter) genes cloned into pUC119, Apr | This study |
| pUCemrAB | 3.9-kb SalI-BamHI fragment containing emrR (regulator), -A (MFP), and -B (multidrug transporter) genes cloned into pUC119, Apr | This study |
| pUCemrD | 2.0-kb HindIII-SacI fragment containing emrD (multidrug transporter) gene cloned into pUC119, Apr | This study |
| pUCyidY | 2.7-kb PstI-BamHI fragment containing yidY (putative transporter) and -Z (putative regulator) genes cloned into pUC119, Apr | This study |
| pUCyieO | 2.2-kb PstI-BamHI fragment containing yieP (hypothetical protein) and -O (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCyjiO | 2.8-kb SphI-BamHI fragment containing yjiO (putative transporter) and -N (hypothetical protein) genes cloned into pUC119, Apr | This study |
| Plasmids carrying MATE-type transporter ORFs | ||
| pUCydhE | 1.6-kb PstI-BamHI fragment containing ydhE (multidrug transporter) gene cloned into pUC119, Apr | This study |
| Plasmids carrying RND-type transporter ORFs | ||
| pUCacrAB | 6.0-kb PstI SmaI fragment containing acrR (regulator), -A (MFP), and -B (multidrug transporter) genes cloned into pUC119, Apr | This study |
| pUCcusA | 9.0-kb SphI-EcoRI fragment containing cusS (formerly ybcZ; sensor), cusR/C/F/B (ylcA/B/C/D: regulator/putative OMF/putative binding protein/putative MFP), and cusA (ybdE: putative metal ion or drug transporter) genes cloned into pUC119, Apr | This study |
| pUCyegMNOB | 14.2-kb SphI-EcoRI fragment containing yegK/L (putative two-component system), yegM/N/O/B (putative MFP/putative RND transporter/putative RND transporter/putative MF transporter), and baeS/R (putative two-component system) genes cloned into pUC119, Apr | This study |
| pUCacrD | 3.5-kb HindIII-SacI fragment containing acrD (drug transporter) gene cloned into pUC119, Apr | This study |
| pUCacrEF | 5.9-kb SphI-SalI fragment containing envR (regulator), acrE/F (MFP/multidrug transporter) genes cloned into pUC119, Apr | This study |
| pUCyhiUV | 4.7-kb PstI-BamHI fragment containing yhiU/V (putative MFP/putative transporter) genes cloned into pUC119, Apr | This study |
| Plasmids carrying SMR-type transporter ORFs | ||
| pUCemrE | 1.2-kb SmaI-BamHI fragment containing emrE (drug transporter) gene cloned into pUC118, Apr | This study |
| pUCydgFE | 1.1-kb SalI-BamHI fragment containing ydgF/E (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCsugE | 1.0-kb PstI-SmaI fragment containing sugE (putative transporter) gene cloned into pUC119, Apr | This study |
| Plasmids carrying ABC-type transporter ORFs | ||
| pUCmdlAB | 4.2-kb SphI-EcoRI fragment containing mdlA/B (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCybjYZ | 3.7-kb PstI-SalI fragment containing ybjY/Z (putative MFP/putative transporter) genes cloned into pUC119, Apr | This study |
| pUCyddA | 2.3-kb SalI-SacI fragment containing yddA (putative transporter) gene cloned into pUC119, Apr | This study |
| pUCyojIH | 4.0-kb SphI-SalI fragment containing yojI/H (putative transporter) genes cloned into pUC119, Apr | This study |
| pUCyhiH | 5.8-kb SphI-BamHI fragment containing yhiI/H (putative regulator/putative transporter) and yhhJ (hypothetical protein) genes cloned into pUC119, Apr | This study |
| Plasmids carrying drug transporter ORFs under control of trc promoter | ||
| MF type | ||
| pTrcHyajR | 1.4-kb NcoI-PstI fragment containing yajR gene cloned into pTrc6His, Apr | This study |
| pTrcHyceE | 1.2-kb NcoI-PstI fragment containing yceE gene cloned into pTrc6His, Apr | This study |
| pTrcHyceL | 1.2-kb NcoI-PstI fragment containing yceL gene cloned into pTrc6His, Apr | This study |
| pTrcHydeA | 1.2-kb NcoI-PstI fragment containing ydeA gene cloned into pTrc6His, Apr | This study |
| pTrcHydeF | 1.2-kb EcoRI-PstI fragment containing ydeF gene cloned into pTrc6His, Apr | This study |
| pTrcHynfM | 1.3-kb NcoI-PstI fragment containing ynfM gene cloned into pTrc6His, Apr | This study |
| pTrcHydhC | 1.2-kb NcoI-SalI fragment containing ydhC gene cloned into pTrc6His, Apr | This study |
| pTrcHydiM | 1.2-kb NcoI-SalI fragment containing ydiM gene cloned into pTrc6His, Apr | This study |
| pTrcHyebQ | 1.5-kb NcoI-SalI fragment containing yebQ gene cloned into pTrc6His, Apr | This study |
| pTrcHyegB | 1.4-kb NcoI-PstI fragment containing yegB gene cloned into pTrc6His, Apr | This study |
| pTrcHbcr | 1.2-kb NcoI-PstI fragment containing bcr gene cloned into pTrc6His, Apr | This study |
| pTrcHyidY | 1.2-kb NcoI-PstI fragment containing yidY gene cloned into pTrc6His, Apr | This study |
| pTrcHyieO | 1.4-kb NcoI-PstI fragment containing yieO gene cloned into pTrc6His, Apr | This study |
| pTrcHyjiO | 1.2-kb NcoI-PstI fragment containing yjiO gene cloned into pTrc6His, Apr | This study |
| RND type | ||
| pTrcHcusAB | 4.4-kb NcoI-XbaI fragment containing cusA and cusB genes cloned into pTrc6His, Apr | This study |
| pTrcHyegN | 3.1-kb NcoI-PstI fragment containing yegN gene cloned into pTrc6His, Apr | This study |
| pTrcHyegO | 3.1-kb EcoRI-PstI fragment containing yegO gene cloned into pTrc6His, Apr | This study |
| pTrcHacrD | 3.1-kb NcoI-SalI fragment containing acrD gene cloned into pTrc6His, Apr | This study |
| pTrcHacrEF | 4.3-kb SacI-PstI fragment containing acrE and acrF genes cloned into pTrc6His, Apr | This study |
| SMR type | ||
| pTrcHydgFE | 0.7-kb EcoRI-PstI fragment containing ydgF and ydgE genes cloned into pTrc6His, Apr | This study |
| pTrcHsugE | 0.5-kb NcoI-PstI fragment containing sugE gene cloned into pTrc6His, Apr | This study |
| ABC type | ||
| pTrcHmdlA | 1.8-kb EcoRI-PstI fragment containing mdlA gene cloned into pTrc6His, Apr | This study |
| pTrcHmdlB | 1.8-kb NcoI-PstI fragment containing mdlB gene cloned into pTrc6His, Apr | This study |
| pTrcHybjZ | 2.0-kb NcoI-PstI fragment containing ybjZ gene cloned into pTrc6His, Apr | This study |
| pTrcHyddA | 1.7-kb NcoI-PstI fragment containing yddA gene cloned into pTrc6His, Apr | This study |
| pTrcHyojI | 1.6-kb NcoI-PstI fragment containing yojI gene cloned into pTrc6His, Apr | This study |
| pTrcHyojH | 1.7-kb EcoRI-PstI fragment containing yojH gene cloned into pTrc6His, Apr | This study |
| pTrcHyhiH | 2.7-kb NcoI-PstI fragment containing yhiH gene cloned into pTrc6His, Apr | This study |
DNA fragments cloned in pUC vectors contained the endogenous promoter regions that were expected to work at the drug resistance measurements without induction.
TABLE 3.
Drug resistance of E. coli KAM3 harboring plasmids carrying putative drug transporter ORFs under control of the trc promotera
| Compound | MIC (μg/ml) without plasmid | MIC (μg/ml) with plasmid carrying ORF for transporter of type:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MF
|
RND
|
SMR (pTrcHydgFE) | |||||||||
| pTrcHyceE | pTrcHyceL | pTrcHbcrb | pTrcHyidY | pTrcHyjiO | pTrcHcusAB | pTrcHyegO | pTrcHacrD | pTrcHacrEF | |||
| Chloramphenicol | 0.39 | 0.39 | 0.39 | 0.39 | 0.78 | 0.78 | 0.39 | 0.39 | 0.39 | 6.25 | 0.39 |
| Tetracycline | 0.39 | 0.39 | 0.39 | 3.13 | 0.39 | 0.39 | 0.39 | 0.39 | 0.78 | 3.13 | 0.39 |
| Minocycline | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 3.13 | 0.20 |
| Erythromycin | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 200 | 3.13 |
| Nalidixic acid | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 1.56 | 1.56 | 3.13 | 1.56 |
| Norfloxacin | 0.025 | 0.025 | 0.05 | 0.025 | 0.025 | 0.05 | 0.025 | 0.05 | 0.05 | 0.10 | 0.025 |
| Enoxacin | 0.05 | 0.05 | 0.10 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.39 | 0.05 |
| Kanamycin | 6.25 | 6.25 | 6.25 | 25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 |
| Vancomycin | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
| Fosfomycin | 1.56 | 6.25 | 1.56 | 6.25 | 1.56 | 1.56 | 3.13 | 3.13 | 3.13 | 1.56 | 3.13 |
| Doxorubicin | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | >200 | 3.13 |
| Novobiocin | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 6.25 | 50 | 1.56 |
| Rifampin | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Trimethoprim | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 100 | 3.13 |
| Acriflavine | 12.5 | 12.5 | 12.5 | 25 | 12.5 | 50 | 12.5 | 12.5 | 12.5 | 400 | 12.5 |
| Crystal violet | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 12.5 | 0.78 |
| Ethidium bromide | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 400 | 12.5 |
| Rhodamine 6G | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 400 | 6.25 |
| Methylviologen | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 200 | 100 |
| TPP | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 200 | 6.25 |
| CCCP | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Benzalkonium | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 50 | 3.13 |
| SDS | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 100 | >400 | >400 | 100 |
| Deoxycholate | 1,250 | 2,500 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 5,000 | >40,000 | >40,000 | 5,000 |
Values in bold differ significantly from the KAM3 control strain value, and 0.1 mM IPTG was added to agar plates. pTrcHyajR, pTrcHydeA, pTrcHydeF, pTrcHynfM, pTrcHydhC, pTrcHydiM, pTrcHyebQ, pTrcHyegB, pTrcHyieO, pTrcHyegN, pTrcHsugE, pTrcHmdlA, pTrcHmdlB, pTrcHybjZ, pTrcHyddA, pTrcHyojI, pTrcHyojH, and pTrcHyhiH did not alter MIC values.
pTrcHbcr conferred bicyclomycin resistance (MIC, 25 μg/ml for KAM3 versus 400 μg/ml for KAM3/pTrcHbcr).
Construction of multicopy plasmid library containing putative drug transporter ORFs.
ORFs assumed to be drug transporter genes were cloned from E. coli W3104 as follows. Chromosomal DNA from E. coli W3104 was isolated as described (46). ORFs were amplified with native promoters and peripheral genes by means of PCR using primers containing the restriction enzyme site that exists in the multicloning sites of the pUC118 and pUC119 vectors. The DNA fragments were digested with restriction enzymes and then ligated into the multicloning sites of pUC118 and pUC119. The fragment sizes, multicloning sites used, and deduced functions of the putative products of ORFs are shown in Table 1. All drug transporter ORFs were arranged to be in the same orientation as the lactose promoter of pUC vectors, while some of the peripheral ORFs were in the reverse orientation. The initial amplification of the recombinant plasmids was done in TG1, and plasmid DNA from five independent colonies of every recombinant was extracted, followed by transformation of KAM3 cells with these plasmids. The resistance to drugs and toxic compounds was measured without induction because the lac promoters in these plasmids were separated by about 300 to 500 bp from the nearest ORFs and the endogenous promoters were expected to work. The nucleotide sequences of the recombinant plasmids were determined by the dideoxy chain termination method (47) using a DNA sequencer (ALF Express; Pharmacia Biotech). Sequence data were analyzed with Genetyx sequence analysis software (Software Development Co.). The SwissProt and GenBank databases were screened for sequence similarities.
Construction of expression plasmid library containing putative drug transporter ORFs.
We constructed the original expression vector pTrc6His as follows. The oligonucleotides 5′-GCATCACCATCACCATCACTA-3′, which codes for hexahistidine, and 5′-AGCTTAGTGATGGTGATGGTGATGCTGCA-3′ were annealed together. The resulting annealed fragment was inserted into the PstI and HindIII sites of the pTrc99A expression vector to produce pTrc6His. Drug transporter ORFs were amplified by PCR using primers containing the restriction enzyme site that exists in the multicloning sites of the pTrc6His vector, and then the DNA fragments were ligated into the vector to produce an expression plasmid library (Table 1). Competent KAM3 cells were transformed with at least five of the constructed plasmids that were extracted from independent colonies, and then the susceptibilities of all transformants to various drugs were measured with induction by 0.1 mM IPTG, and protein expression was detected with antipolyhistidine antibodies.
RESULTS AND DISCUSSION
Cloning and analysis of MF- and MATE-type drug transporter ORFs.
In the MF family, there are 19 ORFs on the chromosomal DNA of E. coli that can be assumed to be drug transporters on the basis of sequence similarities (Table 1). Among these transporters, EmrB, EmrY, YebQ, and YegB possess 14 hydrophobic regions, and Fsr, MdfA (Cmr), YdeA, Bcr, EmrD, YceE, YdeF, YdhC, YidY, YieO, YjiO, YajR, YceL, YnfM, and YdiM possess 12 hydrophobic regions, which may be transmembrane domains, as judged from hydropathy analysis according to Eisenberg et al. (7) (data not shown). All of these transporters possessing 14 hydrophobic regions have the glycine-rich motif (motif C) in transmembrane segment V that is conserved in the MF-type drug transporters (13, 42). Transporters possessing 12 hydrophobic regions have the GXXXX(R/K)XGR(R/K) motif (motif A) in the putative cytoplasmic loop between transmembrane segments II and III that is conserved in the MF-type drug transporters (42, 55). Therefore, it is likely that these transporters are members of the MF family. In the MATE family, only ydhE had been identified as a multidrug resistance (MDR) gene (26).
In these two families, EmrA/B (19), Fsr (8), MdfA (Cmr) (6, 32), Bcr (1), EmrD (29), YjiO (6), and YdhE (26) had been identified as drug exporters, and YdeA (5) had been reported as an exporter of l-arabinose and IPTG, although the transport capabilities of EmrY, YebQ, YegB, YceE, YdeF, YdhC, YidY, YieO, YajR, YceL, YnfM, and YdiM have not been established yet. We amplified these 20 ORF clusters with the endogenous promoters and some peripheral ORFs from the chromosomal DNA of E. coli W3104 by PCR. The PCR fragments were cloned into multicopy pUC vectors (Table 1). The MICs of 24 different compounds for E. coli KAM3 cells harboring these plasmids were measured. We used a broad range of toxic compounds, including representative cationic dyes, antimicrobial agents, antiseptics, anticancer drugs, uncouplers, and detergents that are well translocated by AcrAB or other major drug transporters. E. coli KAM3, which lacks AcrAB, showed hypersensitivity to these compounds (Table 2). The MICs were measured without IPTG so that the ORFs were expected to be expressed under the control of the endogenous promoters.
TABLE 2.
Drug resistance of E. coli cells harboring plasmids carrying putative drug transporter ORFsa
| Compound | MIC (μg/ml) without plasmid | MIC (μg/ml) with plasmid carrying ORF for transporter of type:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MF
|
MATE (pUCydhE) | RND
|
SMR
|
ABC (pUCybjYZ) | ||||||||||||||
| pUCfsrb | pUCmdfA | pUCyceE | pUCbcrc | pUCemrAB | pUCemrD | pUCynfM | pUCyebQ | pUCacrAB | pUCyegMNOBd | pUCacrD | pUCacrEF | pUCyhiUV | pUCemrE | pUCydgFE | ||||
| Chloramphenicol | 0.39 | 0.39 | 6.25 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.78 | 3.13 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 |
| Tetracycline | 0.39 | 0.39 | 0.78 | 0.39 | 1.56 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 3.13 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 | 0.39 |
| Minocycline | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 3.13 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 |
| Erythromycin | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 100 | 3.13 | 3.13 | 3.13 | 25 | 3.13 | 3.13 | 25 |
| Nalidixic acid | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 3.13 | 1.56 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 |
| Norfloxacin | 0.025 | 0.025 | 0.20 | 0.025 | 0.025 | 0.025 | 0.025 | 0.025 | 0.025 | 0.20 | 0.20 | 0.05 | 0.025 | 0.025 | 0.025 | 0.025 | 0.025 | 0.025 |
| Enoxacin | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.39 | 0.39 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
| Kanamycin | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Vancomycin | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
| Fosfomycin | 1.56 | 1.56 | 1.56 | 6.25 | 6.25 | 1.56 | 1.56 | 1.56 | 1.56 | 3.13 | 1.56 | 3.13 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 |
| Doxorubicin | 3.13 | 3.13 | 12.5 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 25 | >200 | 3.13 | 3.13 | 6.25 | 25 | 3.13 | 3.13 | 3.13 |
| Novobiocin | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 100 | 25 | 6.25 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 |
| Rifampin | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Trimethoprim | 3.13 | 6.25 | 12.5 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 0.78 | 12.5 | >100 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 |
| Acriflavine | 12.5 | 12.5 | 100 | 12.5 | 25 | 12.5 | 12.5 | 3.13 | 12.5 | 12.5 | >200 | 12.5 | 12.5 | 100 | 12.5 | 200 | 12.5 | 12.5 |
| Crystal violet | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 12.5 | 1.56 | 1.56 | 1.56 | 3.13 | 3.13 | 1.56 | 1.56 |
| Ethidium bromide | 12.5 | 12.5 | 50 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 25 | >400 | 12.5 | 12.5 | 50 | 50 | 100 | 12.5 | 12.5 |
| Rhodamine 6G | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 6.25 | 400 | 6.25 | 6.25 | 25 | 100 | 6.25 | 6.25 | 6.25 |
| Methylviologen | 100 | 100 | 100 | 100 | 100 | 200 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 200 | 100 | 100 |
| TPP | 6.25 | 6.25 | 25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 200 | 400 | 6.25 | 6.25 | 6.25 | 25 | 6.25 | 6.25 | 6.25 |
| CCCP | 6.25 | 12.5 | 6.25 | 6.25 | 6.25 | 50 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Benzalkonium | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 3.13 | 6.25 | 3.13 | 3.13 | 6.25 | 100 | 6.25 | 3.13 | 3.13 | 6.25 | 6.25 | 3.13 | 3.13 |
| SDS | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 50 | 50 | >400 | 200 | >400 | 100 | 200 | 50 | 100 | 50 |
| Deoxycholate | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 40,000 | 1,250 | 1,250 | 1,250 | 40,000 | >40,000 | >40,000 | >40,000 | 5,000 | 5,000 | 1,250 | 5,000 | 1,250 |
Values in bold differ significantly from the KAM3 control strain value. MICs were measured without IPTG, so that the ORFs were expected to be expressed under the control of the endogenous promoters. Data are not shown for pUCemrKY, which carries emrK/Y and evgS/A genes. This plasmid conferred resistance to doxorubicin (MIC, 100 μg/ml), rhodamine 6G (100 μg/ml), benzalkonium (25 μg/ml), erythromycin (12.5 μg/ml), crystal violet (6.25 μg/ml), TPP (25 μg/ml), deoxycholate (5,000 μg/ml), fosfomycin (3.13 μg/ml), novobiocin (3.13 μg/ml), acriflavine (25 μg/ml), ethidium bromide (25 μg/ml), methylviologen (200 μg/ml), and SDS (100 μg/ml). Multidrug resistance conferred by pUCemrKY is due to overexpression of evgA, emrK/Y genes confer deoxycholate resistance (33). pUCyajR, pUCyceL, pUCydeA, pUCydeF, pUCydhC, pUCydiM, pUCyidY, pUCyieO, pUCyjiO, pUCcusA, pUCsugE, pUCmdlAB, pUCyddA, pUCyojIH, and pUCyhiH did not alter MIC values.
pUCfsr conferred fosmidomycin resistance (MIC, 0.39 μg/ml for KAM3 versus 12.5 μg/ml for KAM3/pUCfsr).
pUCbcr conferred bicyclomycin resistance (MIC, 25 μg/ml for KAM3 versus 400 μg/ml for KAM3/pUCbcr).
pUCyegMNOB carries both RND- (yegM/N/O) and MF-type (yegB) ORFs. Resistance conferred by pUCyegMNOB is due to overexpression of RND genes (unpublished data).
In the MF family, nine different plasmids (pUCemrAB, pUCfsr, pUCmdfA, pUCbcr, pUCemrD, pUCydhE, pUCemrKY, pUCyegMNOB, and pUCyceE) conferred drug resistance on E. coli KAM3 cells (Table 2). pUCemrAB conferred resistance to deoxycholate (32-fold the wild-type level), CCCP (8-fold), rhodamine 6G (2-fold), methylviologen (2-fold), and SDS (2-fold). Thus, EmrB, which is a putative transmembrane protein, seems to be a multidrug transporter, as reported by Lomovskaya and Lewis (19). emrA encodes a protein containing a single hydrophobic domain and a C-terminal hydrophilic domain. This gene is needed for emrB-modulated drug resistance (19). The gene fsr was believed to code for resistance to a single drug, fosmidomycin (8). pUCfsr indeed increased resistance to fosmidomycin (32-fold), but also unexpectedly increased slightly (2-fold) the MIC of trimethoprim and CCCP (Table 2). pUCmdfA conferred resistance to chloramphenicol (16-fold), norfloxacin (8-fold), acriflavine (8-fold), doxorubicin (4-fold), trimethoprim (4-fold), ethidium bromide (4-fold), TPP (4-fold), and tetracycline (2-fold), as reported by Edgar and Bibi (6). pUCbcr conferred resistance to tetracycline (4-fold), fosfomycin (4-fold), kanamycin (2-fold), and acriflavine (2-fold). Since the insert region of pUCbcr possesses two ORFs (yejD and bcr), we cloned the bcr gene alone into the pTrc6His expression vector (pTrcHbcr) under the control of the trc promoter and investigated the drug resistance phenotype in the presence of IPTG. Protein expression was detected with an antipolyhistidine antibody (Fig. 1). pTrcHbcr exhibited a drug resistance pattern comparable to that of pUCbcr (Table 3). These two plasmids also conferred bicyclomycin resistance (16-fold) (Tables 2 and 3), as reported previously (1). Although bcr had been reported to codes for resistance to a single drug, bicyclomycin (1), this gene seems to confer a moderate increase in resistance to some other compounds. pUCemrD conferred resistance to SDS (2-fold) and benzalkonium (2-fold). Naroditskaya et al. (29) reported that the emrD gene confers resistance to some uncouplers. This gene also conferred resistance to detergents. pUCydhE conferred increased resistance to TPP (32-fold), deoxycholate (32-fold), norfloxacin (8-fold), enoxacin (8-fold), doxorubicin (8-fold), trimethoprim (4-fold), chloramphenicol (2-fold), fosfomycin (2-fold), ethidium bromide (2-fold), and benzalkonium (2-fold), indicating that this gene confers resistance to a far broader range of compounds, including toxic compounds, than reported by Morita et al. (26). pUCemrKY conferred resistance to doxorubicin (32-fold), rhodamine 6G (16-fold), benzalkonium (8-fold), erythromycin (4-fold), crystal violet (4-fold), TPP (4-fold), deoxycholate (4-fold), fosfomycin (2-fold), novobiocin (2-fold), acriflavine (2-fold), ethidium bromide (2-fold), methylviologen (2-fold), and SDS (2-fold). The insert region of this plasmid carries four ORFs (evgS, evgA, emrK, and emrY) (Table 1). In our previous study (33), we found that overexpression of evgA, which is a response regulator of a two-component system, confers multidrug resistance by stimulating the expression of several MDR transporters. Although the expression of emrK/Y is also regulated by evgA (15), emrK/Y only conferred deoxycholate resistance (33). pUCyegMNOB conferred resistance to deoxycholate (32-fold), novobiocin (16-fold), SDS (4-fold), nalidixic acid (2-fold), norfloxacin (2-fold), and fosfomycin (2-fold). This plasmid contains eight ORFs in its insert region (Table 1), and this region contains both MF- (yegB) and RND-type (yegN/O) transporter ORFs.
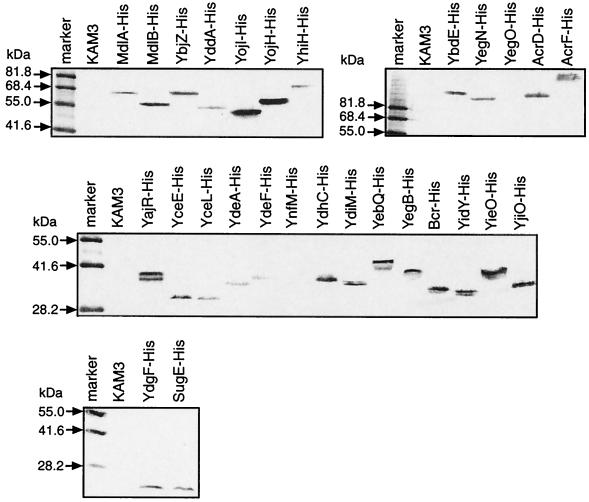
FIG. 1.
Expression of putative drug transporter ORFs. Twenty-eight putative drug transporter ORFs were cloned into pTrc6His expression vectors to produce hexahistidine-tagged proteins under control of the trc (trp/lac hybrid) promoter. E. coli KAM3 cells harboring the constructed plasmids were cultured in 2×YT medium in the presence of 0.1 mM IPTG for induction of protein expression. The cells were harvested and then disrupted by sonication. Total cell proteins were separated on an SDS-polyacrylamide gel, and protein expression was detected by Western blot analysis with an antipolyhistidine antibody.
We cloned yegB into an expression vector (pTrcHyegB) and investigated the resulting drug resistance phenotype. yegB expression could be detected (Fig. 1), but this gene did not confer resistance to any of the compounds listed in Table 3. Identification of the drug resistance gene in this locus is discussed later. pUCyceE conferred fosfomycin resistance (4-fold) on E. coli (Table 2). We also cloned yceE into an expression vector (pTrcHyceE). Expression of yceE could be detected (Fig. 1), and pTrcHyceE conferred resistance to fosfomycin (4-fold) and deoxycholate (2-fold) in the presence of IPTG (Table 3). Deoxycholate resistance of pUCyceE might not be detectable because the expression level of yceE was lower than that of pTrcHyceE. yceE seems to be a novel resistance gene.
E. coli cells harboring multicopy plasmids carrying either yajR, yceL, ydeA, ydeF, ynfM, ydhC, ydiM, yebQ, yidY, yieO, or yjiO did not show any resistance to the compounds tested in this study (Table 2). Since there is a possibility that these ORFs are not expressed from their native promoters, we cloned these ORFs into the pTrc6His expression vector. The expression of all these ORFs except ynfM could be detected (Fig. 1). We found again that yceL, yidY, and yjiO conferred drug resistance (Table 3). yceL conferred resistance to norfloxacin (2-fold) and enoxacin (2-fold). yidY conferred chloramphenicol resistance (2-fold). yjiO conferred resistance to acriflavine (4-fold), chloramphenicol (2-fold), norfloxacin (2-fold), ethidium bromide (2-fold), and TPP (2-fold), indicating that this gene confers resistance to a broader range of compounds than previously reported (6). ydeA has been reported to be an l-arabinose and IPTG exporter (5), but this gene did not confer resistance to any of the compounds tested in this study. pUCynfM and pUCyebQ conferred hypersensitivity to acriflavine and trimethoprim, respectively (Table 2). However, when ynfM or yebQ was expressed alone from an expression vector, these genes did not confer the hypersensitivity phenotype (Table 3). In the case of the pUC plasmid library, these genes were cloned simultaneously with putative regulatory gene ynfL or kdgR, respectively (Table 1). Thus, it seems that overexpression of ynfL or kdgR in the induction of acriflavine or trimethoprim would be deleterious to cell growth
In summary, we found two novel multidrug resistance determinants (yceE and yceL) and one chloramphenicol resistance gene (yidY). Two drug-specific determinants, fsr and bcr, were revealed to be multidrug resistance ones. In addition, emrD, yjiO, and ydhE conferred broader resistance than hitherto believed.
Cloning and analysis of RND-type drug transporter ORFs.
In the RND family, there are seven ORFs on the chromosomal DNA of E. coli that can be assumed to be drug transporters on the basis of sequence similarities. AcrB, AcrD, AcrF, YhiV, CusA (formerly YbdE), YegN, and YegO possess 12 hydrophobic regions, which may be transmembrane domains, as judged from hydropathy analysis according to Eisenberg et al. (7) (data not shown). AcrD, AcrF, YhiV, CusA, YegN, and YegO exhibit sequence similarity to AcrB at 76, 84, 79, 41, 47, and 48% similarity and 66, 77, 71, 20, 28, and 28% identity, respectively, in the amino acid sequences.
Of these seven ORFs, AcrA/B (21), AcrD (43), and AcrE/F (16, 22; J. Xu, M. L. Nilles, and K. P. Bertrand, Abstr. 93rd Gen. Meet. Am. Soc. Microbiol. 1993, abstr. K-169, p. 290, 1993) had been identified as drug exporters. It was reported that yhiU/V shows homology to acrA/B (20, 22) and yhiU/V confers octane resistance on E. coli (52), but deletion of this gene pair does not increase the susceptibility of E. coli (49). Recently, the ybdE gene was renamed cusA (27) and has been shown to be involved in the efflux of copper (10). The transport capabilities of YegN and YegO have not been reported yet.
We cloned these ORFs with some peripheral ORFs into multicopy plasmids (Table 1) and investigated their drug resistance phenotypes (Table 2). Among them, pUCacrAB, pUCacrD, pUCacrEF, pUCyhiUV, and pUCyegMNOB conferred drug resistance. pUCacrAB conferred increased resistance to doxorubicin (>64-fold), novobiocin (64-fold), rhodamine 6G (64-fold), TPP (64-fold), trimethoprim (>32-fold), ethidium bromide (>32-fold), deoxycholate (>32-fold), erythromycin (32-fold), benzalkonium (32-fold), acriflavine (>16-fold), minocycline (16-fold), SDS (>8-fold), chloramphenicol (8-fold), tetracycline (8-fold), norfloxacin (8-fold), enoxacin (8-fold), crystal violet (8-fold), nalidixic acid (8-fold), and rifampin (2-fold). This broad specificity is the same as that reported previously (20, 22, 34, 51). pUCacrD conferred resistance to deoxycholate (>32-fold), SDS (>8-fold), novobiocin (4-fold), and kanamycin (2-fold). When acrD was cloned into an expression vector, it slightly increased (2-fold) the MIC of tetracycline, nalidixic acid, norfloxacin, and fosfomycin in addition to deoxycholate (>32-fold), SDS (>8-fold), novobiocin (4-fold), and kanamycin (2-fold) (Table 3). AcrD expression is shown in Fig. 1. Although the acrD gene was reported to mediate an aminoglycoside resistance (43), this gene confers resistance to some other compounds in addition to an aminoglycoside. pUCacrEF conferred resistance to acriflavine (8-fold), ethidium bromide (4-fold), rhodamine 6G (4-fold), deoxycholate (4-fold), doxorubicin (2-fold), and SDS (2-fold). The insert region of pUCacrEF possesses three genes (envR, acrE, and acrF) (Table 1). Since envR is a regulatory gene for acrE/F (envC/D) (2, 17, 21), we cloned the acrE/F gene pair into an expression vector (pTrcHacrEF). Expression of acrF was detected, as shown in Fig. 1, and pTrcHacrEF conferred far broader resistance to doxorubicin (>64-fold), erythromycin (64-fold), rhodamine 6G (64-fold), trimethoprim (>32-fold), deoxycholate (>32-fold), novobiocin (32-fold), acriflavine (32-fold), ethidium bromide (32-fold), TPP (32-fold), chloramphenicol (16-fold), minocycline (16-fold), crystal violet (16-fold), benzalkonium (16-fold), SDS (>8-fold), tetracycline (8-fold), enoxacin (8-fold), nalidixic acid (4-fold), norfloxacin (4-fold), and methylviologen (2-fold) (Table 3) than pUCacrEF. This is because envR acts as a repressor of acrEF. pUCyhiUV conferred resistance to rhodamine 6G (16-fold), erythromycin (8-fold), doxorubicin (8-fold), ethidium bromide (4-fold), TPP (4-fold), SDS (4-fold), deoxycholate (4-fold), crystal violet (2-fold), and benzalkonium (2-fold), indicating that YhiV is a multidrug resistance determinant. YhiU seems to be a membrane fusion protein (MFP) like AcrA. Although pUCcusA did not show any resistance to the compounds tested in this study (Table 2), when cusA (ybdE) was cloned into an expression vector with the putative MFP ORF cusB (ylcD) (pTrcHcusAB), this plasmid conferred fosfomycin resistance (2-fold) in the presence of IPTG (Table 3). pUCyegMNOB conferred resistance to deoxycholate (32-fold), novobiocin (16-fold), SDS (4-fold), nalidixic acid (2-fold), norfloxacin (2-fold), and fosfomycin (2-fold) as described above. This plasmid contains eight ORFs, including three putative drug transporter ORFs, yeg-N (RND), -O (RND), and-B (MF) (Table 1). We separately cloned these three transporter ORFs into an expression vector (pTrcHyeg-N, -O, or -B) and then investigated their drug resistance phenotypes in the presence of IPTG (Table 3). The expression of YegN and YegB was detectable, while YegO was not detectable with an antipolyhistidine antibody (Fig. 1), possibly because the C terminus of YegO undergoes processing and the histidine tag may be removed. Out of three expression plasmids, only pTrcHyegO conferred drug resistance to deoxycholate (4-fold), nalidixic acid (2-fold), norfloxacin (2-fold), fosfomycin (2-fold), and SDS (2-fold), but lacked novobiocin resistance. Since YegM exhibits 47% similarity and 28% identity to the AcrA membrane fusion protein and the other ORFs (yegK/L and baeS/R) included in pUCyegMNOB are not transporter ORFs, there is a possibility that a multicomplex(es) formation (for example, YegM/N/O) may act as a transporter. When the yegMNO genes were cloned together into vector pUC, this plasmid conferred a drug resistance phenotype comparable to that of pUCyegMNOB, but the plasmid carrying the yegM/N genes did not confer resistance. (S. Nagakubo, K. Nishino and A. Yamaguchi, unpublished data). Thus, resistance conferred by pUCyegMNOB seems to be due to overexpression of MFP and RND genes. Partially, overexpression of yegO is responsible for the drug resistance in this complex.
Cloning and analysis of SMR-type drug transporter ORFs.
There are three ORFs that can be assumed to encode SMR-type drug exporters on the basis of sequence similarities. All three of these ORFs (emrE, ydgF/E, and sugE) encode putative membrane proteins having four putative hydrophobic transmembrane regions. EmrE, YdgF/E, and SugE consist of 110, 109/121, and 155 amino acid residues, respectively. YdgF/E and SugE exhibit similarity to EmrE (56%/53% and 54% similarity and 35%/32% and 33% identity at the amino acid level). YdgF/E is considered a two-component-type SMR transporter (14).
Among them, only EmrE had been identified as a multidrug exporter, although the transport capabilities of YdgF/E and SugE have not been reported yet. We cloned these ORFs into multicopy vectors (Table 1), and investigated the drug resistance (Table 2). pUCemrE and pUCydgFE conferred drug resistance on E. coli KAM3. The emrE gene conferred resistance to acriflavine (16-fold), ethidium bromide (8-fold), crystal violet (2-fold), methylviologen (2-fold), and benzalkonium (2-fold), the resistance phenotype being the same as that reported by Yerushalmi et al. (57). pUCydgFE conferred resistance to deoxycholate (4-fold) and SDS (2-fold). We also cloned ydgFE into an expression vector (pTrcHydgFE). YdgE expression is shown in Fig. 1. pTrcHydgFE conferred broader resistance to deoxycholate (4-fold), nalidixic acid (2-fold), fosfomycin (2-fold), and SDS (2-fold) (Table 3) than pUCydgFE. This is because the expression level of ydgFE with the expression vector was higher than that with the multicopy vector. These results indicate that ydgFE is a novel drug resistance determinant. sugE did not confer resistance to any of the compounds tested in this study (Tables 2 and 3), although the expression of sugE was detected (Fig. 1).
Cloning and analysis of ABC-type drug transporter ORFs.
There are seven ORFs that can be assumed to be genes encoding ABC-type drug exporters on the basis of sequence similarities. MdlA, MdlB, YddA, YojI, YojH, and YhiH each possess six hydrophobic putative transmembrane regions, and YbjZ possesses four hydrophobic regions. Among them, only YhiH has two nuleotide-binding domains, and the others have a single nucleotide-binding domain. None of these has yet been reported to have transport capability. We cloned all of these ORFs with some peripheral ORFs into pUC vectors (Table 1), and the resulting drug resistance was investigated (Table 2). Only pUCybjYZ conferred increased resistance on E. coli KAM3. This plasmid confers erythromycin-specific resistance (8-fold). However, YbjZ has no sequence homology to MsrA (44), which is an ABC-type macrolide-specific exporter in gram-positive cocci, except for the nucleotide-binding domain. We also cloned seven putative ABC-type ORFs into expression vectors (pTrcH-mdlA, -mdlB, -ybjZ, -yddA, -yojI, yojH, and -yhiH). None of them conferred resistance to any of the compounds listed in Table 3, although their expression was detected (Fig. 1). Overexpression of ybjY alone also did not confer drug resistance. It seems that ybjY and ybjZ make a complex for the drug resistance phenotype, and this gene pair exhibited resistance against macrolides composed of 14- and 15-membered lactones, such as clarithromycin, oleandomycin, and azithromycin, in addition to erythromycin (17a). Thus, ybjY/Z is a novel macrolide resistance determinant. This is the first experimentally identified ABC-type drug resistance gene in gram-negative bacteria.
In this study, the 37 putative drug exporter genes in E. coli were all cloned into multicopy vectors and expression vectors, and the resistance phenotypes against 26 structurally different compounds were investigated. As a result, we found that 20 genes (11 MF, 2 SMR, 6 RND, and 1 ABC) conferred drug resistance. Among them, we identified seven novel drug resistance determinants, yceE, yceL, yidY, ydgFE, yegO, cusA (ybdE), and ybjYZ (3 MF, 1 SMR, 2 RND, and 1 ABC), their drug resistance capabilities having not yet been reported. When yegMNO were expressed together, this complex gave broader resistance spectra than yegO alone. The ybjZ gene is the first case of an experimentally identified ABC drug exporter gene in gram-negative bacteria. In addition, the fsr, bcr, yjiO, ydhE, acrD, and yhiUV genes gave broader resistance spectra than previously reported.
The remaining 17 of the 37 putative drug transporter ORFs did not confer resistance to the compounds tested in this study on E. coli. The possible reasons are as follows. (i) These ORFs might confer resistance to compounds other than those tested in this study. (ii) These ORFs might need complex formation with other ORFs for drug resistance. (iii) These ORFs might be unrelated to drug resistance. Recently, Sulavik et al. reported antibiotic susceptibility profiles of E. coli strains lacking multidrug efflux pump genes (49). They reported that deletion of three known MDR pump genes (acrAB, mdfA, and emrE) and three OMF genes (tolC, yjcP, and yohG) resulted in strains with increased susceptibility to some compounds. However, they reported that deletion of other known and predicted MDR pump genes, including yhiUV, yegMNO, and ybjYZ, which were identified as drug resistance determinants in our study, did not increase susceptibility to any compounds. Thus, the method of overexpression analysis in this study is useful for identifying drug resistance factors underlying chromosomal DNA. The drug exporter ORF expression library established in this study is a genetic resource for bacterial drug resistance, and we believe that the library should be useful for the future development of chemotherapy.
ACKNOWLEDGMENTS
We thank T. Tsuchiya and Y. Morita of Okayama University for providing us with the E. coli KAM3 strain.
K. Nishino is supported by a research fellowship from the Japan Society for the Promotion of Science for Young Scientists. This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis H, van Veen H W, Poolman B, Driessen A J, Konings W N. Mechanisms of multidrug transporters. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown M H, Paulsen I T, Skurray R A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 5.Carole S, Pichoff S, Bouch J P. Escherichia coli gene ydeA encodes a major facilitator pump which exports L-arabinose and isopropyl-beta-d-thiogalactopyranoside. J Bacteriol. 1999;181:5123–5125. doi: 10.1128/jb.181.16.5123-5125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. . (Erratum, 179:5654.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 8.Fujisaki S, Ohnuma S, Horiuchi T, Takahashi I, Tsukui S, Nishimura Y, Nishino T, Kitabatake M, Inokuchi H. Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene. 1996;175:83–87. doi: 10.1016/0378-1119(96)00128-x. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg M, Pribyl T, Juhnke S, Nies D H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 10.Grass G, Rensing C, Munson G P, Lam D L, Outten F W, O'Halloran T V. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grinius L, Dreguniene G, Goldberg E B, Liao C H, Projan S J. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid. 1992;27:119–129. doi: 10.1016/0147-619x(92)90012-y. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Iwaki S, Tamura N, Kimura-Someya T, Nada S, Yamaguchi A. Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a permeability barrier in the middle of a transmembrane water-filled channel. J Biol Chem. 2000;275:22704–22712. doi: 10.1074/jbc.m910354199. [DOI] [PubMed] [Google Scholar]
- 14.Jack D L, Storms M L, Tchieu J H, Paulsen I T, Saier M H., Jr A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J Bacteriol. 2000;182:2311–2313. doi: 10.1128/jb.182.8.2311-2313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato A, Ohnishi H, Yamamoto K, Furuta E, Tanabe H, Utsumi R. Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci Biotechnol Biochem. 2000;64:1203–1209. doi: 10.1271/bbb.64.1203. [DOI] [PubMed] [Google Scholar]
- 16.Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett. 1999;179:345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x. [DOI] [PubMed] [Google Scholar]
- 17.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 17a.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183:5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Littlejohn T G, DiBerardino D, Messerotti L J, Spiers S J, Skurray R A. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene. 1991;101:59–66. doi: 10.1016/0378-1119(91)90224-y. [DOI] [PubMed] [Google Scholar]
- 19.Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 23.Masaoka Y, Ueno Y, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J Bacteriol. 2000;182:2307–2310. doi: 10.1128/jb.182.8.2307-2310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurry L, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli Proc. Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J Bacteriol. 2000;182:6694–6697. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein of Vibrio parahaemolyticus, and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munson G P, Lam D L, Outten F W, O'Halloran T V. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muth T R, Schuldiner S. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 2000;19:234–240. doi: 10.1093/emboj/19.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naroditskaya V, Schlosser M J, Fang N Y, Lewis K. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem Biophys Res Commun. 1993;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 30.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsen I W, Bakke I, Vader A, Olsvik O, El-Gewely M R. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol. 1996;178:3188–3193. doi: 10.1128/jb.178.11.3188-3193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishino K, Yamaguchi A. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters J. Bacteriol. 2001;183:1455–1458. doi: 10.1128/JB.183.4.1455-1458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen I T, Littlejohn T G, Radstrom P, Sundstrom L, Skold O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulsen I T, Nguyen L, Sliwinski M K, Rabus R, Saier M H. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen I T, Sliwinski M K, Nelissen B, Goffeau A, Saier M H., Jr Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998;430:116–125. doi: 10.1016/s0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen I T, Sliwinski M K, Saier M H., Jr Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 39.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob Agents Chemother. 2000;44:2595–2599. doi: 10.1128/aac.44.10.2595-2599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 42.Putman M, van Veen H W, Konings W N. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg E Y, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross J I, Eady E A, Cove J H, Cunliffe W J, Baumberg S, Wootton J C. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 45.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PAO1:an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 49.Sulavik M C, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw K J, Miller G H, Hare R, Shimer G. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother. 2001;45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor J W, Ott J, Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985;13:8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukagoshi N, Aono R. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol. 2000;182:4803–4810. doi: 10.1128/jb.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Veen H W, Konings W N. Drug efflux proteins in multidrug resistant bacteria. Biol Chem. 1997;378:769–777. [PubMed] [Google Scholar]
- 54.van Veen H W, Venema K, Bolhuis H, Oussenko I, Kok J, Poolman B, Driessen A J, Konings W N. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc Natl Acad Sci USA. 1996;93:10668–10672. doi: 10.1073/pnas.93.20.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi A, Kimura T, Someya Y, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10: the structural resemblance and functional difference in the role of the duplicated sequence motif between hydrophobic segments 2 and 3 and segments 8 and 9. J Biol Chem. 1993;268:6496–6504. [PubMed] [Google Scholar]
- 56.Yamamoto T, Tanaka M, Nohara C, Fukunaga Y, Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981;145:808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zgurskaya H I, Nikaido H. Bypassing te periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]