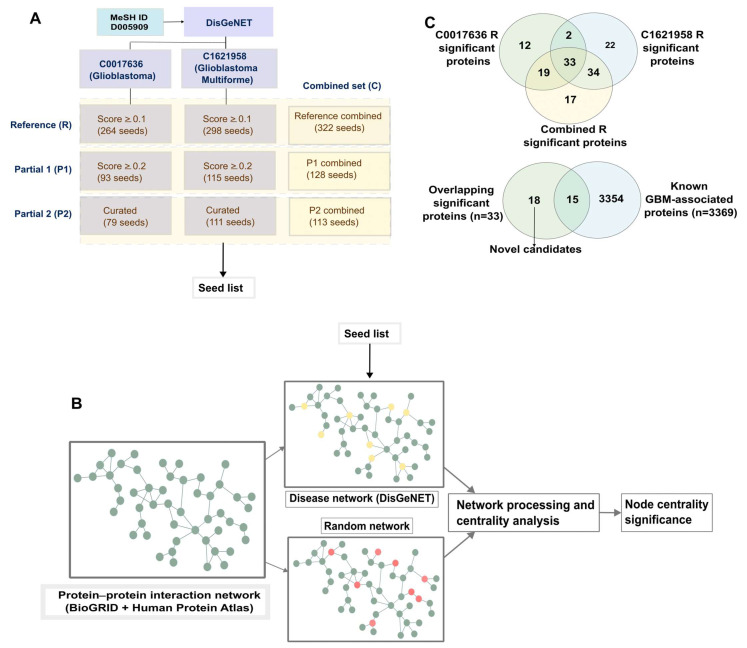
Figure 1.
(A) Disease-associated proteins (DAPs) were obtained from DisGeNET. Two terms corresponding to glioblastoma and glioblastoma multiforme were used to obtain seed lists of various sizes based on different criteria. A third set of combined seed lists was also created. R: Reference (R) sets; P1: sets of DAPs with score ≥ 0.2; P2: sets of DAPs with score ≥ 0.1. (B) For each of the seed lists, the procedure for centrality analysis was followed—mapping seeds to PPI (yellow-DAPs, red-randomly selected nodes), creating disease-specific and degree-stratified background random networks, centrality analysis and processing, to obtain background-corrected node centralities and significance. (C) Overlapping significant proteins obtained from the analysis of the networks of the two categories were obtained as the most consistent candidates.