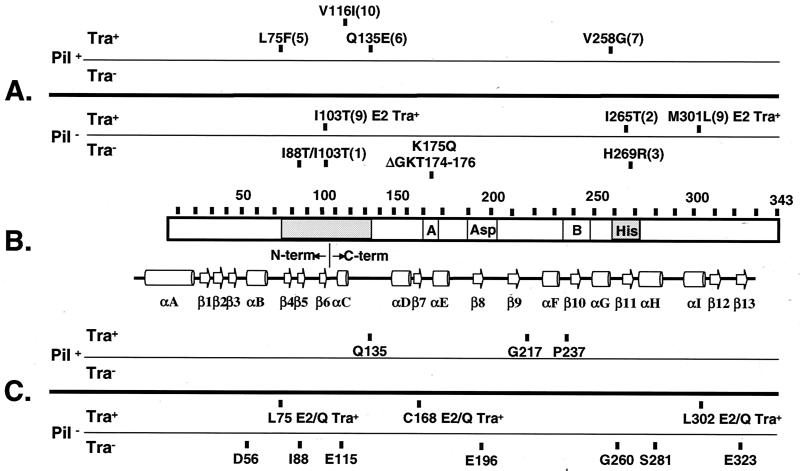
FIG. 7.
Positions of VirB11 mutations grouped according to their effects on pilus production and substrate transfer. (A) Substitution mutations with allele numbers in parantheses as defined by Zhou et al. (69). Phenotypic descriptions are provided for mutations of special interest. Note that all virB11/virB11∗ merodiploid strains expressing the dominant alleles exhibit a T-pilus+ IncQ plasmid Tra+ T-DNA Tra-deficient phenotype. (B) VirB11 (343 residues) with conserved Walker A and B domains and the Asp and His boxes denoted. The shading identifies the two regions of VirB11 in which dominant mutations were predominantly located. Below the VirB11 representation is the HP0525 secondary structure with β-sheets and α-helices as presented by Yeo et al. (67). The junction between the N- and C-terminal domains, shown by the HP0525 crystal structure to assemble as independent hexameric rings, is indicated. (C) i4 mutations with insertion sites indicated.