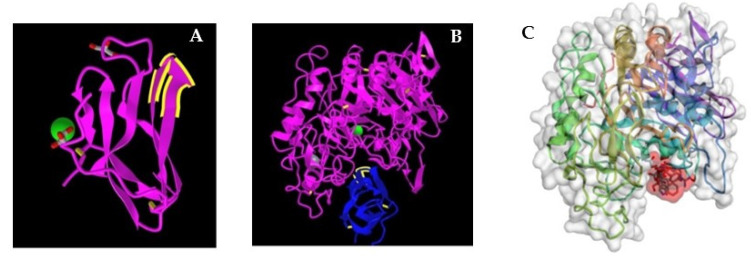
Figure 1.
Structure of the complex of α-amylase with tendamistat. (A) The crystal structure of tendamistat (PDB accession no. 1OK0). The loop structure that binds to the glycan binding site of α-amylase is highlighted. (B) The crystal structure of the complex with tendamistat bound in the glycan binding site of porcine α-amylase (accession no. 1BVN). (C) Model of the 6-mer loop structure of tendamistat (shaded in red) bound to porcine α-amylase (accession no. 1PIF) generated in silico with CABS-Dock (RMSD = 3.561 Å). The predicted binding energy, ΔG′ = −56 to −60 kJ/mol, is nearly the value calculated from Keq.