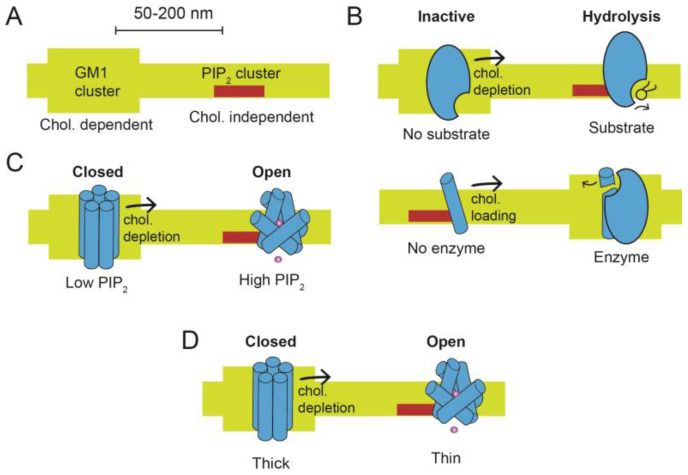
Figure 1.
Cholesterol-regulated protein (CRP) activation by cholesterol depletion. (A) Two lipid domains are shown. On the left, saturated GM1 lipids are thick and form a cholesterol (chol.) dependent location. On the right, phosphatidylinositol 4,5 bisphosphate (PIP2) lipids are clustered and form cholesterol-independent locations. The two locations are separated by 50–200 nm in a cell membrane. (B–D) CRP proteins move in response to cholesterol depletion. In high cholesterol, proteins reside primarily in the GM1 location. In low cholesterol, they shift to or near the PIP2 location. The shift to a new location produces at least three mechanisms of activation. In (B), CRP activation is shown by substrate presentation. (Top) An enzyme (blue shading) is inactive when sequestered in a GM1 lipid cluster, away from its substrate. When cholesterol is low, the GM1 domain is disrupted, and the enzyme moves (black arrow) to the PIP2 location, where it has access to its substrate. A generic lipid substrate is shown (black stick structure), but the substrate could also be a protein. (Bottom) A protein substrate (blue rod) is shown moving in response to cholesterol loading (black arrow) while the hydrolytic enzyme remains stationary. In (C), CRP activation is shown by lipid gating. An ion channel (blue shading) is localized to GM1 clusters, where it is held inactive from a lack of PIP2. Disruption of GM1 lipids (black arrow) allows the channel to move to PIP2 clusters where PIP2 concentration is high. The binding of PIP2 causes a conformational change in the transmembrane domain that opens the channel. In (D), CRP activation is shown by membrane thickness. An ion channel (blue shading) is sequestered into thick lipids (GM1 lipids). The thickness drives an inactive state. When the channel moves from GM1 lipids, the membrane thins, causing the helices to change conformation and open the channel.