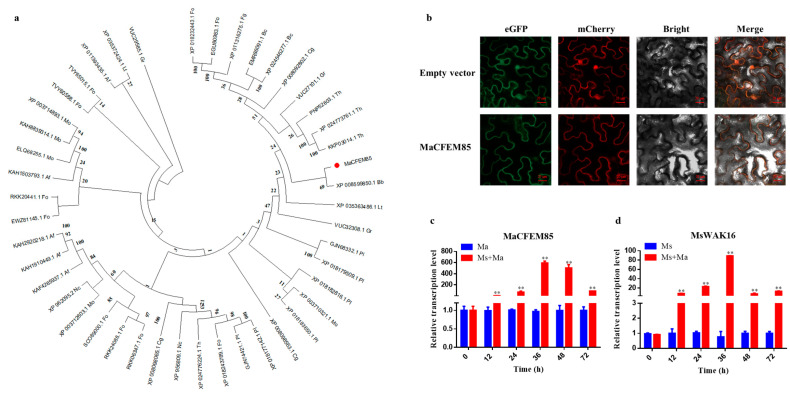
Figure 1.
Phylogenetic analysis, subcellular localization, and expression patterns of MaCFEM85 and MsWAK16. (a) Neighbor-joining phylogenetic tree showing the relationships between MaCFEM85 and CFEM proteins in other fungi. The phylogenic tree was constructed using MEGA X. Mo, Magnaporthe oryzae; Bc, Botrytis cinerea; Fg, Fusarium graminearum; Cg, Colletotrichum graminicola; Fo, Fusarium oxysporum; Nc, Neurospora crassa; Af, Aspergillus fumigatus; Lt, Lasiodiplodia theobromae; Gr, Gliocladium roseum; Th, Trichoderma harzianum; Bb, Beauveria bassiana; Pl, Paecilomyces lilacinus. (b) Subcellular localization of MaCFEM85 proteins. The MaCFEM85–GFP fusion genes were co-expressed with the MaCFEM85-mCherry fusion genes in N. benthamiana leaves. The control vector carried eGFP and mcherry driven by the 35S promotor. Photos were taken at 30 h after infiltration with laser scanning confocal microscopy of eGFP (488 nm excitation and 507 nm emission), mcherry (587 nm excitation and 610 nm emission), bright field microscopy, and merged confocal and bright field. (c,d) qRT-PCR analysis of MaCFEM85 and MsWAK16 expression in co-incubated M. anisopliae and M. sativa. Ma, M. anisopliae; Ms, M. sativa; Ma+Ms, co-incubated Ma and Ms. Error bars show the standard deviation of three biological replicates. ** p < 0.01 (one-way ANOVA with Duncan’s multiple-range t-test).