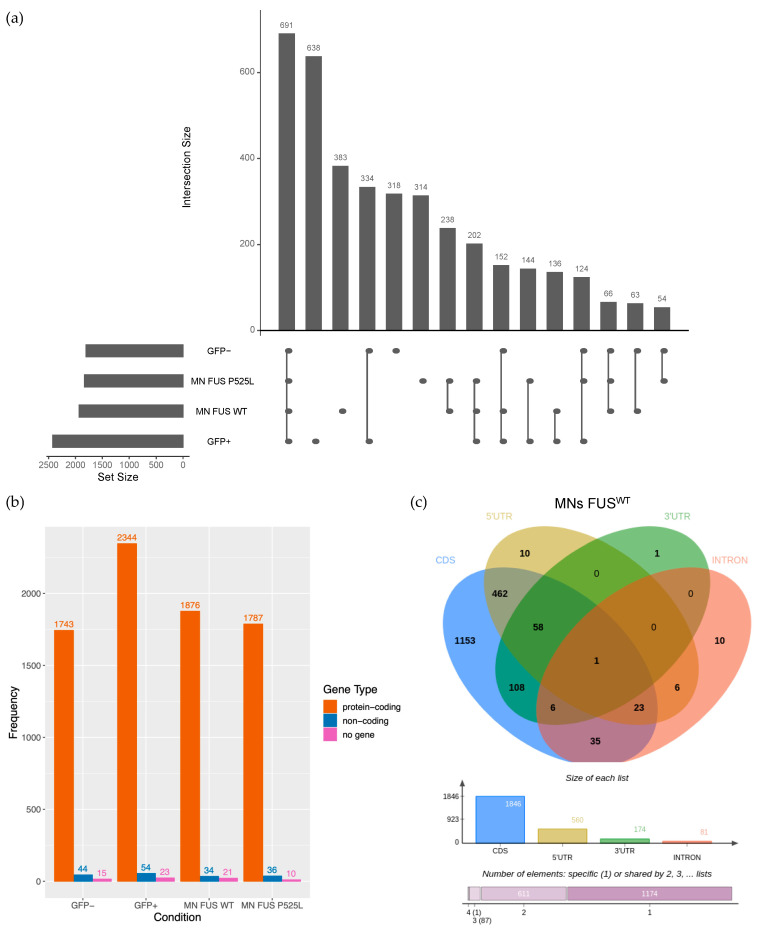
Figure 1.
Distribution of circRNAs in iPSCs-derived MN precursors and mature MNs. (a) UpSet plot showing the number of circRNAs identified (average CPM > 1) in GFP-, GFP+, FUSWT, and FUSP525L cell types, as well as the overlap between such conditions. (b) Bar chart showing, for each cell type, the number of circRNAs hosted by protein-coding genes, non-coding genes, and intergenic regions (no gene). (c) Venn diagram and bar charts showing the protein-coding gene regions occupied by the circRNAs detected in MN FUSWT samples. CDS: coding sequence; 5′UTR: 5′ untranslated region; 3′UTR: 3′ untranslated region; INTRON: intronic region.