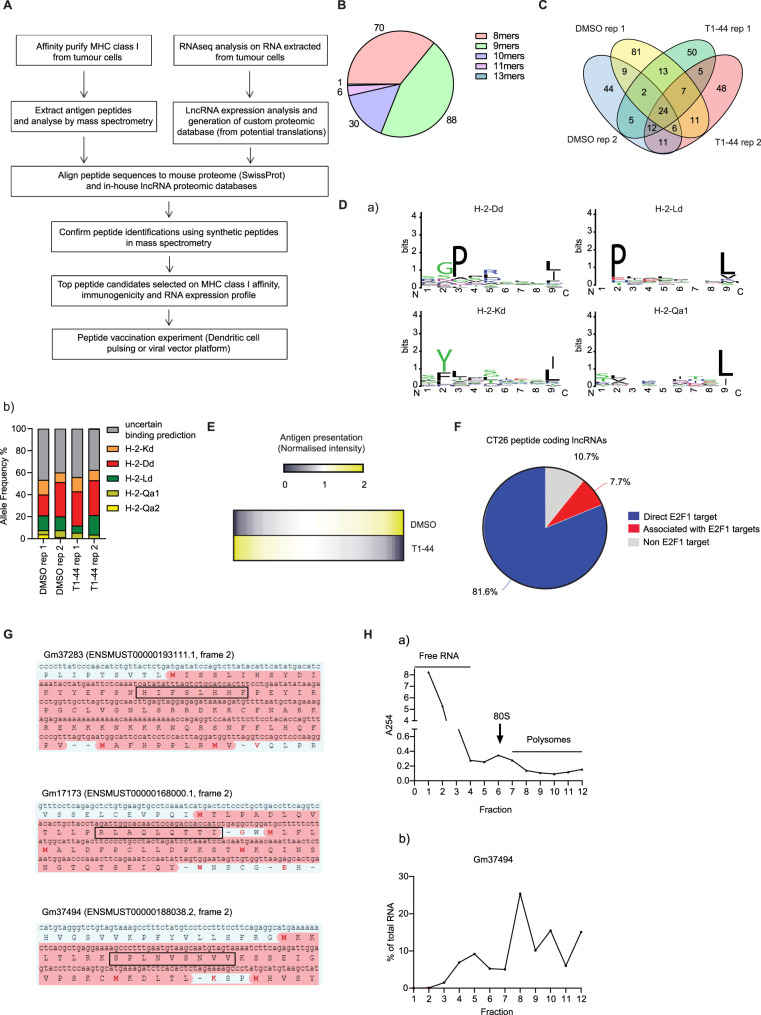
Fig. 2. Immunopeptidomics analysis of CT26 cells.
A Workflow of the immunopeptidomics platform. B Peptide length of each murine lncRNA-derived peptide is displayed as pie chart. 195 quantifiable peptides in total (pooled from two independent experiments) were detected (data derived from an immunopeptidomics experiment performed in biological independent replicate. C Immunopeptidomics analysis in T1-44 treated (1 µM for 72 h) or DMSO treated CT26 cells. The experiment was performed in biological replicate (rep1, rep2). Indicated is the overlap of MHC-bound lncRNA-derived peptides identified from the qualitative immunopeptidomics analysis. 328 peptides were detected. D (a) Sequence logos of amino acid conservation in lncRNA-derived peptides for each of the indicated MHC alleles; (b) Predicted MHC allele frequency for identified lncRNA-derived peptides are displayed, represented as a percentage of total. E Heatmap of lncRNA derived peptide abundance from the quantitative immunopeptidomics analysis (195 peptides) (both up- and down-regulated in T1-44 treatment with respect to DMSO). Relative abundance values were converted by normalisation to the mean. Yellow colour represents up-regulation, whilst blue colour represents down-regulation. Ivory colour represents no change in abundance. n = 2 independent experiments (each with two technical replicates); F The percentage of lncRNA genes giving rise to MHC class I bound peptides that score as potential direct E2F1 target genes or are associated with other potential E2F1 target genes. The analysis was performed on all unique peptide coding lncRNA genes identified in our immunopeptidomics analyses. G Part of the Gm37283, Gm17173, and Gm37494 lncRNA transcripts are displayed, with the predicted ORF (shown in red) giving rise to the identified MHC class I bound peptide (boxed in black). Potential start methionine residues are highlighted in red text. H (a) Example polysome profiling assay from CT26 cells, indicating total RNA quantity detected in each collected fraction (by absorbance reading at 254 nm). (b) Polysome profiling assay for Gm37494 is displayed. Data are presented as percentage of total RNA in each fraction; n = 3 independent experiments (each with three technical replicates).