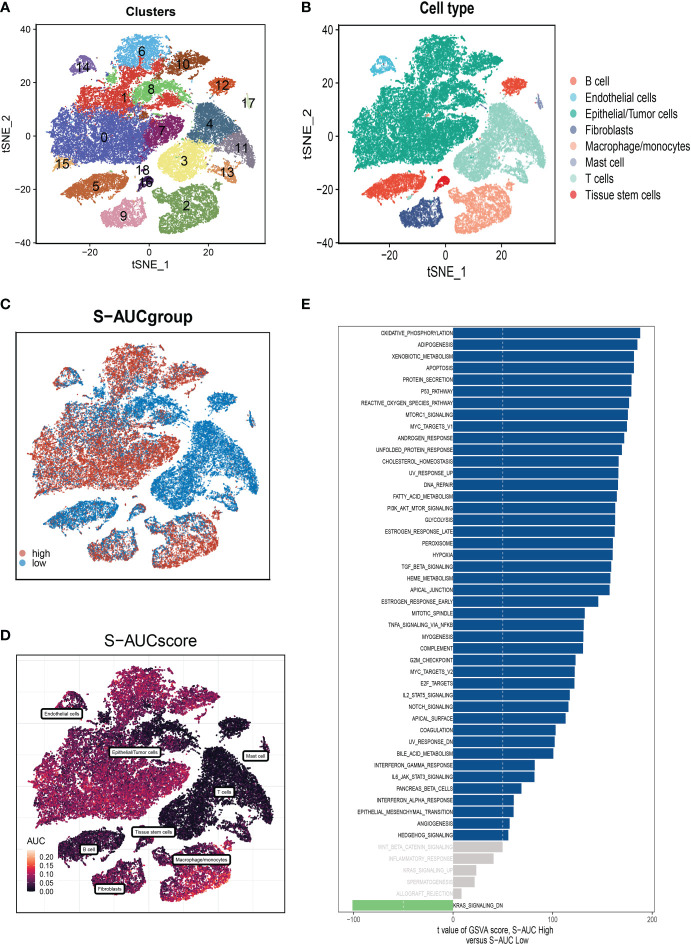
Figure 2.
Annotation of cell subsets and identification of differentially expressed genes. (A) The results of the dimension reduction cluster analysis are shown in the tSNE diagram. (B) Cells were annotated into 8 different types of cells. (C, D) All cells were scored according to sphingolipid-associated genes (SRG) and were divided into high and low groups. (E) Analysis of differentially expressed genes between high and low groups.