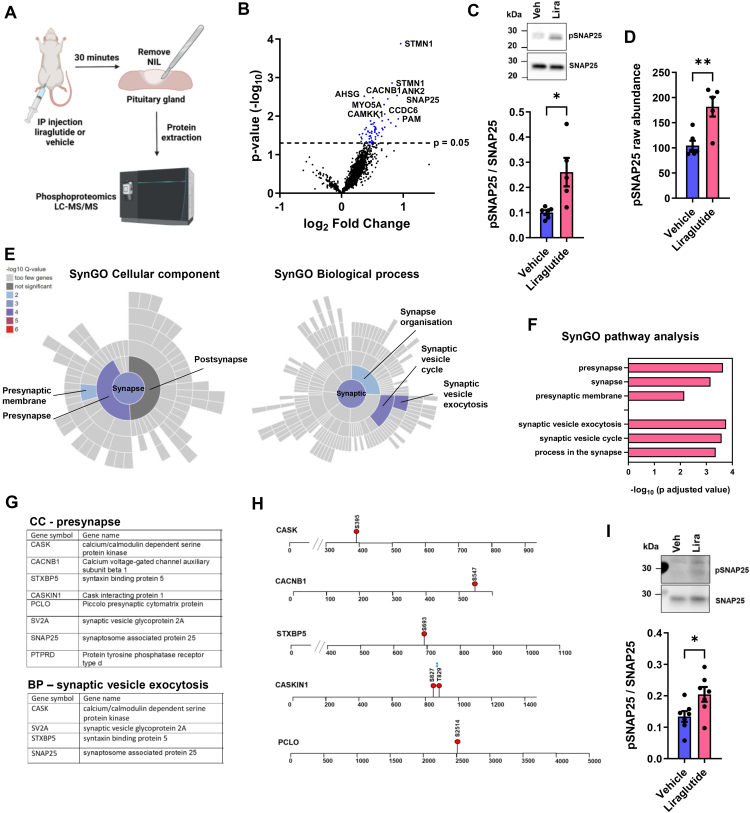
Figure 8.
The liraglutide treated NIL phosphoproteome reveals phosphosite changes to components of the SNARE complex that facilitate vesicular exocytosis. A, outline of the experimental protocol. B, volcano plot of phosphoprotein changes in rat NILs after liraglutide treatment compared to vehicle controls. Selected protein names have been highlighted. C, immunoblot of pSNAP25 and SNAP25 immunoreactive bands in the NIL after liraglutide treatment compared to vehicle controls. Densitometry analysis of immunoreactive bands for pSNAP25 and SNAP25. D, phospho protein raw abundance of pSNAP25 in the NIL according to LC-MS/MS between control and 30-minute liraglutide treated rats. E, analysis of phophoproteomics hyperphosphorylated proteins by SynGO. Figures displaying colour coded enriched terms by Q value for the ontology terms Cellular Component and Biological Process. All level terms identified have been labelled. F, bar chart showing SynGO enriched terms for Cellular Component and Biological Process displayed a -log10 p adjusted value. G, table of hyperphosphorylated proteins containing the overrepresented terms in SynGo ontologies Cellular Component and Biological Process. H, mapping of selected phosphosites undergoing hyperphosphorylation in response to 30 min liraglutide treatment compared to vehicle controls. Blues stars indicate the identification of a novel phosphosite for CASKIN1. I, immunoblots of pSNAP25 and SNAP25 immunoreactive bands in the NIL after liraglutide treatment ex vivo compared to vehicle controls. Densitometry analysis of immunoreactive bands for pSNAP25 and SNAP25 from ex vivo NILs. Values are means + SEM of n = 5 animals per group or n = 7 half pituitaries per group. ∗p ≤ 0.05, ∗∗p ≤ 0.01.