Fig. 1.
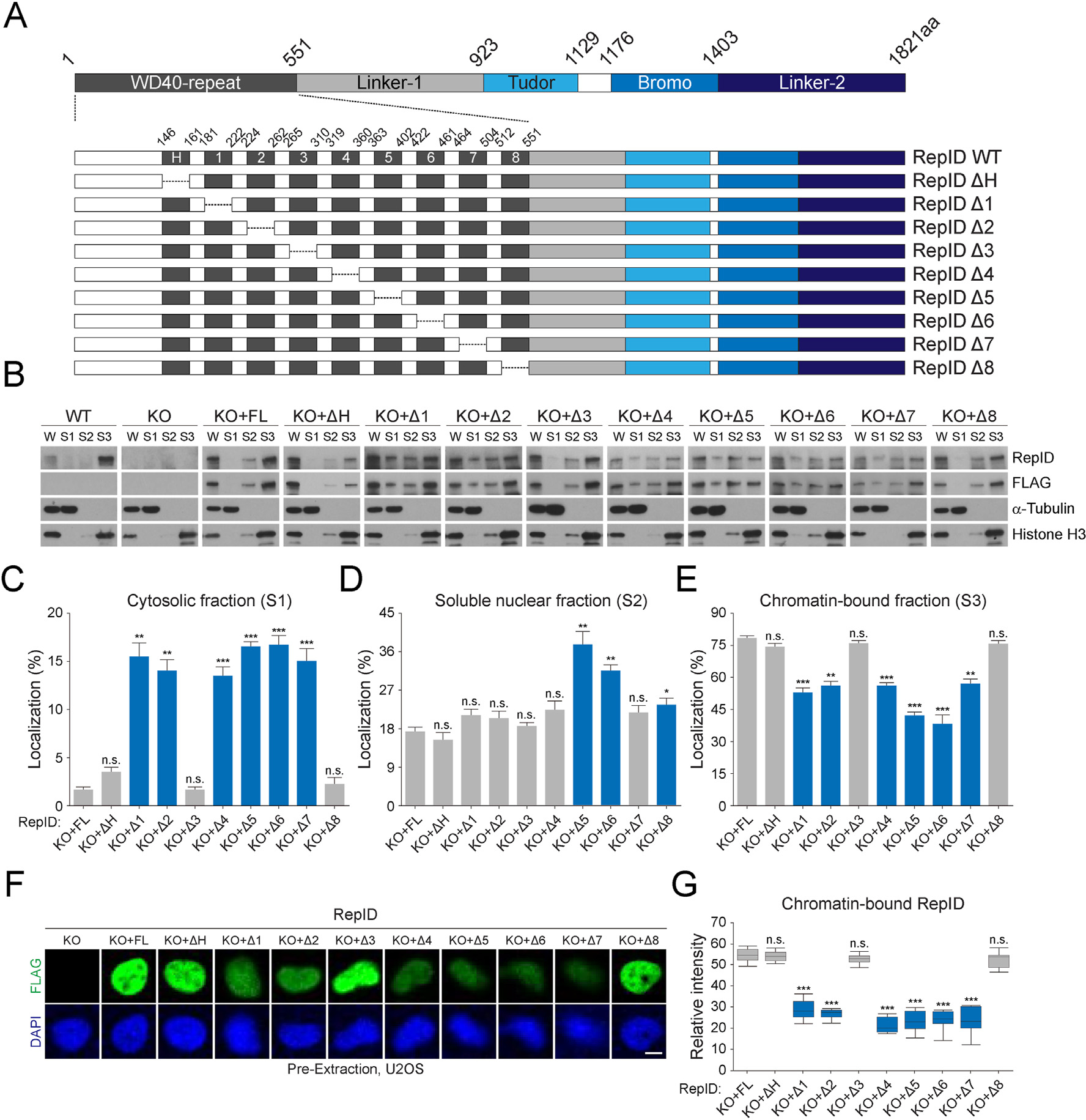
RepID WD40 domain contributes to its chromatin binding. (A) RepID domains and WD40 deletion mutants. WD40 domain consisting of 9 exons including H-box and deletion constructs of each exon is represented with amino acid composition. (B) Levels of RepID full-length (FL) or its deletion mutants from U2OS cells transfected with RepID fragments are indicated. W, whole cell lysates; S1, cytosolic fractions; S2, soluble nuclear fractions; S3, chromatin-bound fractions. Histone H3 and α-tubulin were used as loading controls and fraction markers. (C–E) Quantification of relative RepID signals in cytosolic (C), soluble nuclear (D), and chromatin-bound fractions (E) analyzed as shown in B after normalization with respect to histone H3 or α-tubulin signal intensities, followed by normalization with intensities in whole cell lysates in each cell lines. Error bars represent standard deviations from three independent experiments (*p value < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant, Student’s t-test). (F) U2OS cells were pre-extracted and chromatin-bound RepID using FLAG antibodies (green) and DNA content (DAPI; blue) were detected using immunofluorescence analysis. Scale bar indicates 10 μm. (G) Quantification of relative intensity of each RepID mutant and p-values were calculated using a two-tailed t-test (***p value < 0.001, n.s., not significant, n = 10). Significant changes are indicated as blue bars.
