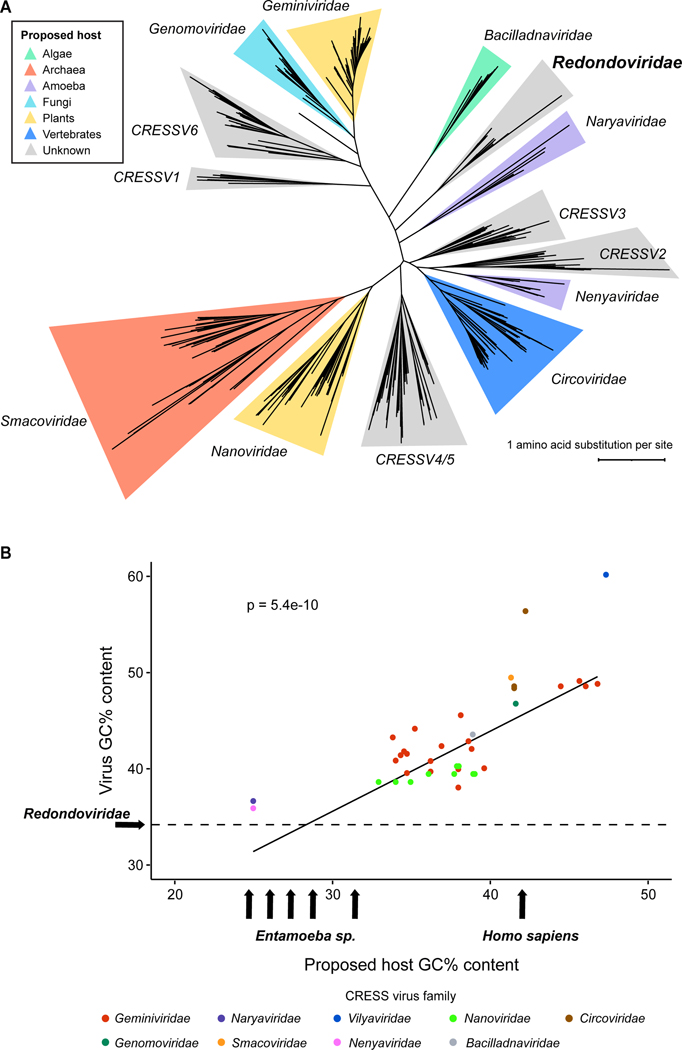
Figure 1.
Investigating the host cell supporting redondovirus replication. (A) Phylogenetic maximum-likelihood tree of Rep amino acid sequences from 441 members of the Cressdnaviricota phylum (Table S1). Rep sequences were aligned with MUSCLE53, followed by tree construction using RaxML54 and visualization using iTOL55. Clade coloring denotes the proposed host of the respective CRESS virus family. (B) Comparative analysis of GC content (% GC) of representative CRESS viruses and their hosts. Virus GC content positively correlates with host GC content (R2 = 0.63, Pearson’s Correlation Test p-value = 5.442e-10, n = 41). The Entamoeba arrows denote (from left to right): E. dispar (24.1%), E. histolytica (24.95%), E. nuttalli (25.1%), E. moshkovskii (26.5%), and E. invadens (30.3%). The dotted horizontal line represents the median GC content of Redondoviridae (34.25%). Figures adapted from Kinsella and coworkers 13.