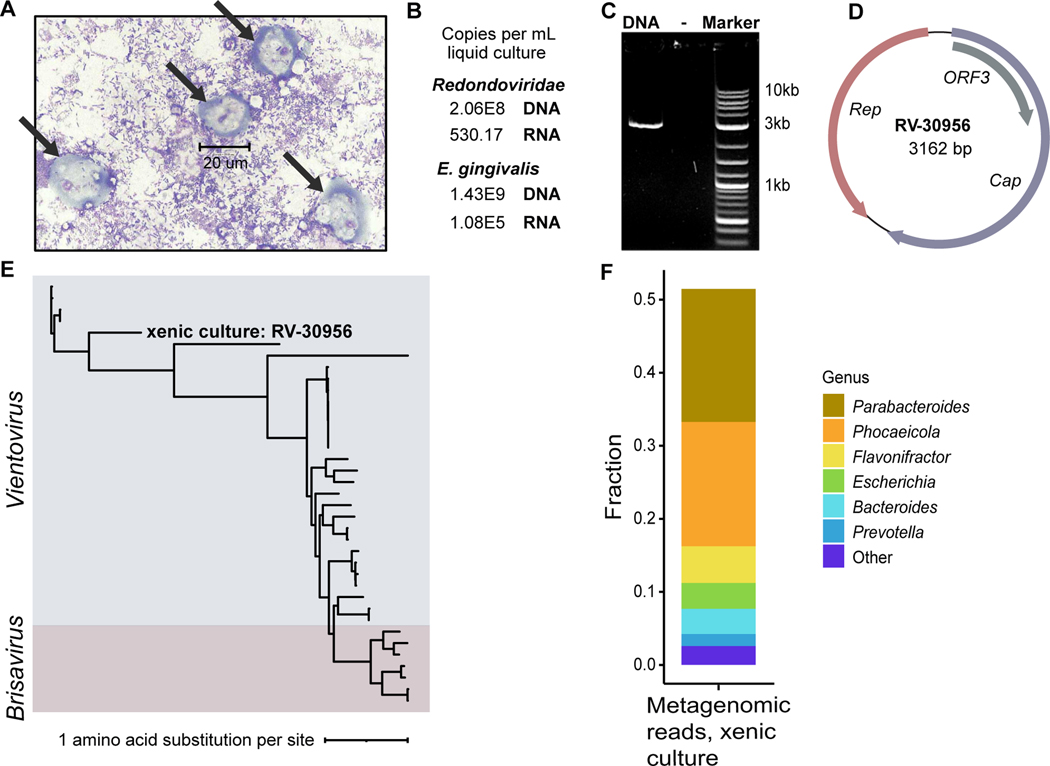
Figure 4.
Detection of redondovirus DNA and RNA in a xenic E. gingivalis culture. (A) Image of the xenic E. gingivalis culture (ATCC-30956) at 100X magnification stained using Kwik-Diff. Scale bar measures a cell (~20 um) that is morphologically consistent a E. gingivalis trophozoite (arrows). No cells with morphology expected for human cells were observed. (B) Detection of redondovirus and E. gingivalis DNA and RNA in the E. gingivalis culture by qPCR and RT-qPCR. (C) Agarose gel electrophoresis showing redondovirus genomic DNA amplification product of the expected size (~3 kb), generated by PCR of DNA from the xenic culture with “back-to-back” PCR primers targeting the circular redondovirus genome. (D) Genome map of the Vientovirus sequenced from the xenic E. gingivalis culture (RV-30956). The largest ORF (violet) encodes the putative capsid (Cap) protein, the second largest (salmon) encodes the Replication-associated (Rep) protein, and the third (grey) encodes a protein of unknown function (ORF3). (E) The RV-30956 sequence placed in a Redondoviridae phylogeny. Rep amino acid sequences from 37 redondoviruses were aligned with MUSCLE53. The tree was built using PhyML with branch support determined by approximate likelihood ratio test56 and visualized using iTOL55. (F) Metagenomic sequence analysis of the E. gingivalis xenic culture. Taxa were identified using Kraken257 and BLASTn58. Only results with reads that could be assigned taxonomically are shown.