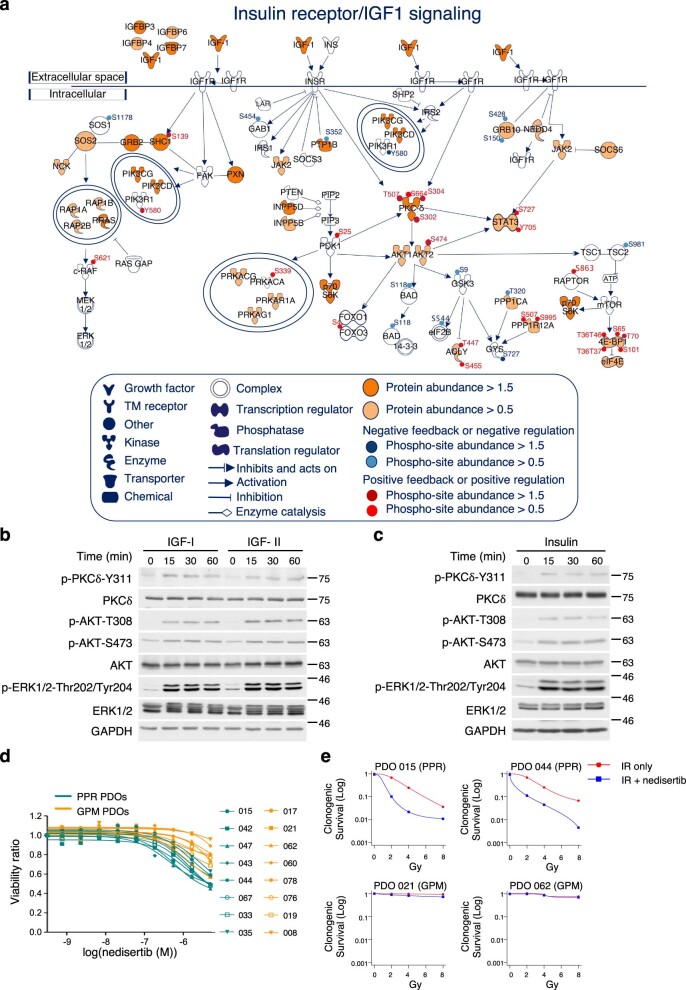
Extended Data Fig. 6. Global and phospho-proteomics events in insulin receptor/IGF-PKCδ pathway in GPM GBM and enrichment of DDR and RS phospho-proteins as a specific feature of PPR GBM.
a, Signaling network highlighting the molecules and proteins involved in IGF-I/insulin signaling of GPM GBM tumors. Orange or red scale indicates the MWW score derived from the proteomic or phosphosite ranked list of GPM tumors when compared to the others, respectively (two-sided MWW test, n = 85 GBM samples). Molecules in white are proteins not profiled or whose abundance was not significantly higher in GPM when compared to the other subtypes. b-c, Western blot analysis of GPM PDO cells incubated with b, IGF-I (10 ng/ml), IGF-II (10 ng/ml) and c, insulin (100 ng/ml) for the indicated times using the indicated antibodies. GAPDH is shown as a loading control. Each experiment was repeated independently 2 times with similar results. d, Viability curves of n = 8 PPR PDOs each derived from an independent patient and n = 8 GPM PDOs, each derived from an independent patient treated with increasing concentration of Nedisertib. Data are mean ± s.d. of n = 4 technical replicates for each PDO from one representative experiment. Experiments were repeated 2 times with similar results. e, Quantification of clonogenic assay of 2 PPR PDOs (PDO 015 and PDO 044, top panels) each derived from an independent patient and 2 GPM PDOs (PDO 021 and PDO 062, bottom panels) each derived from an independent patient treated with IR or IR plus Nedisertib (1667nM). Data are mean of n = 3 technical replicates from one representative experiment. Experiments were repeated 2 times with similar results.