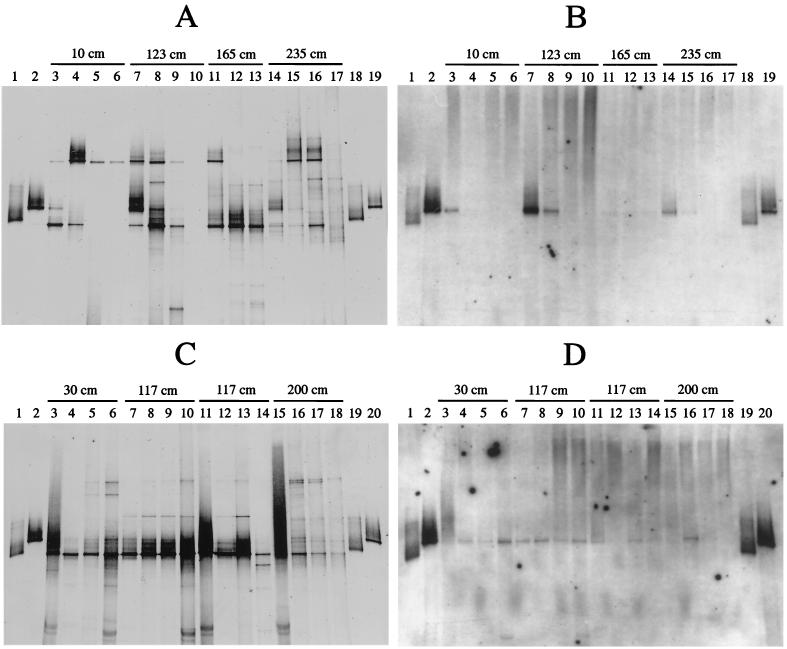
FIG. 2.
Hybridization analysis of DGGE profiles of 16S rDNA fragments obtained with primers specific for Bacteria and template DNA from environmental samples taken along a transect from the shallow-water hydrothermal vent system off Milos and Thiomicrospira isolates obtained from the same location. (A) DGGE pattern of samples taken in June 1996. Lanes: 1 and 18, isolate Milos T-1; 2 and 19, isolate Milos T-2; 3 to 6, samples taken at 10 cm from the vent center (lane 3, surface; lane 4, 0 to 10 mm deep; lane 5, 10 to 20 mm deep; lane 6, 20 to 30 mm deep); 7 to 10, equivalent samples taken at 123 cm from the vent center; 11 to 13, samples taken at 165 cm from the vent center (lane 11, 0 to 10 mm deep; lane 12, 10 to 20 mm deep; lane 13, 20 to 30 mm deep); 14 to 17, equivalent samples taken at 235 cm to those taken at 10 cm. (B) Hybridization analysis of the pattern in panel A with a Thiomicrospira-specific digoxigenin-labeled probe. (C) DGGE pattern of samples taken in September 1996. Lanes: 1 and 19, isolate Milos T-1; 2 and 20, isolate Milos T-2; 3 to 6, samples taken at 30 cm from the vent center (lane 3, surface; lane 4, 0 to 5 mm depth; lane 5, 8 to 13 mm depth; lane 6, 16 to 26 mm depth); 7 to 10, equivalent samples taken at 117 cm from the vent center (sediment core number 1); 11 to 14, equivalent samples taken at 117 cm from the vent center (sediment core number 2); 15 to 18, equivalent samples taken at 200 cm from the vent center. (D) Hybridization analysis of the pattern in panel C with the Thiomicrospira-specific probe.