FIGURE 7.
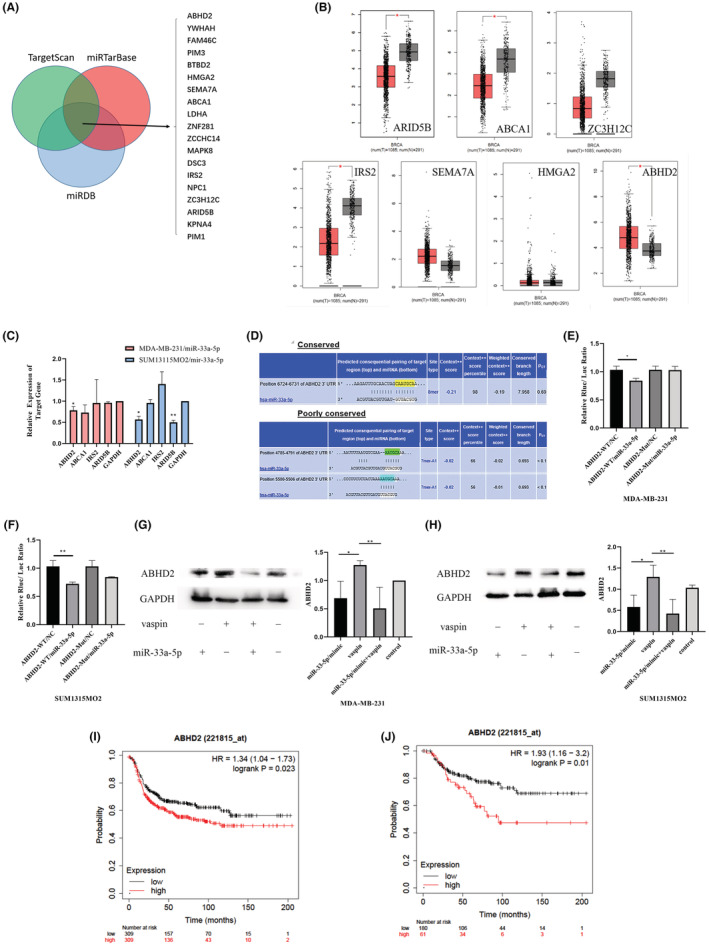
(A) Used Venn diagram to analyze miR‐33a‐5p downstream target genes in miRDB, miRTarBase and TargetScan databases. (B) GEPIA database shows the differential expression ARID5B, ABCA1, ZC3H12C, IRS2, SEMA7A, HMGA2, and ABHD2 in breast cancer. (C) RT‐qPCR results showed differential expression of ABHD2, ABCA1, ARID5B and IRS2 in miR‐33a‐5p/mimic or NC‐transfected MDA‐MB‐231 and SUM1315M02 cells. (D) The TargetScan software predicts that there is a potential binding site for miR‐33a‐5p on the 3'UTR of the ABHD2 gene. (E, F) MDA‐MB‐231 cells and SUM1315MO2 cells were co‐transfected with a wild‐type or mutant 3′‐UTR sequence of ABHD2 and the miR33a‐5p mimic or NC, and the luciferase activity was measured at 48 h and analyzed. (G, H) Western blot experiment to determine the expression and analysis results of the ABHD2 gene in MDA‐MB‐231 cells and SUM1315MO2 cells miR‐33a‐5p/mimic, vaspin, vaspin+miR‐33a‐5p/mimic and the control group. (I) The Kaplan–Meier Plotter database was used to estimate PFS with low (n = 309) or high (n = 309) levels of ABHD2 in patients with triple‐negative breast cancer. (J) The Kaplan–Meier Plotter database was applied to estimate the OS of TNBC patients with low (n = 180) or high (n = 61) levels of ABHD2. Experimental data were performed in triplicates. *p<0.05, **p<0.01
