Figure 3.
Efficient and precise reframing with mRNA in hiPCs
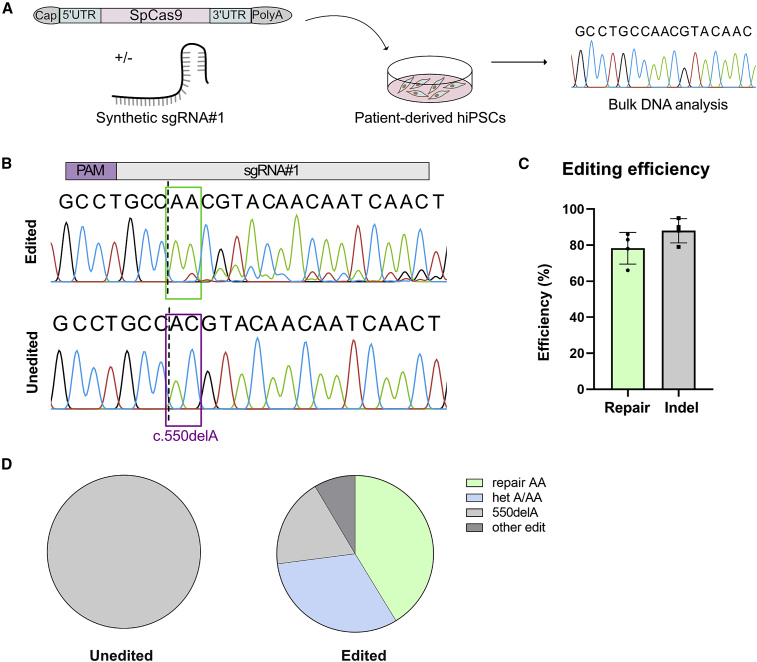
(A) Schematic overview of the experiment. Patient-derived hiPSCs were transfected with sgRNA no. 1 and NLS-SpCas9-NLS mRNA. After expansion cells were processed for bulk DNA analysis. (B) Representative Sanger sequencing result for edited (top) compared with unedited (bottom) sample. Binding site of sgRNA is indicated on top, the purple square marks the position of the mutation, the dotted line represents the cutting site of SpCas9. sgRNA no. 1 leads to a frameshift by insertion of one adenine (green box). (C) Predicted editing efficiency of three biological repeats. The editing efficiency was predicted using ICE tool. In gray the indel frequency is quoted, green shows the repair by +1 insertion. (D) Verification of predicted editing efficiency by analysis of single hiPSC colonies. Single colonies were picked and the DNA sequence of CAPN3 was analyzed with Sanger sequencing. For unedited samples 144 colonies and for edited samples 336 colonies in three biological repeats were analyzed.