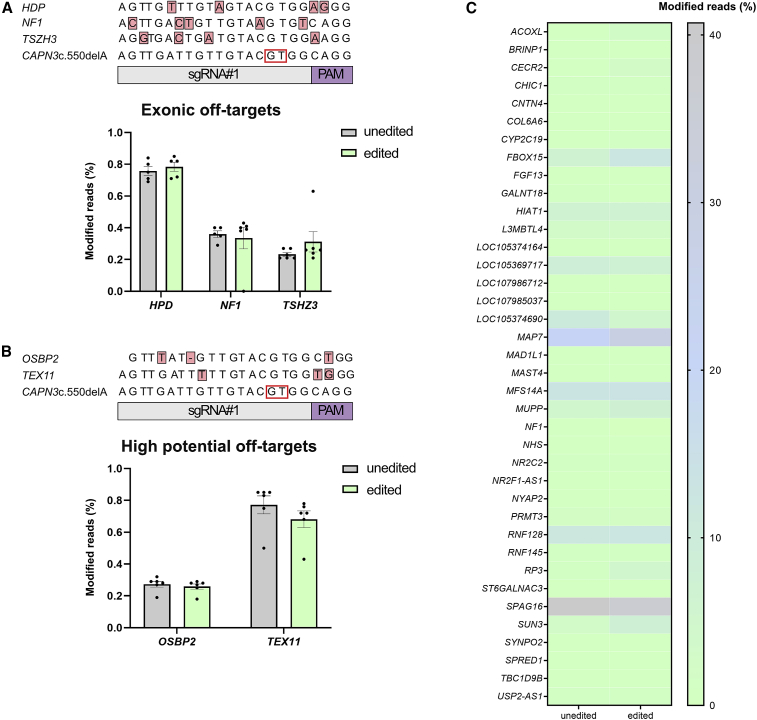
Figure 5.
Off-target analysis
(A) Modification of the predicted exonic off-targets. Percentage of modified reads within all reads at each locus after analysis with amplicon sequencing is plotted. No differences between edited and unedited control can be detected. Two biological repeats for patients 1–3 were analyzed. (B) Modification of the two off-targets predicted by CrispRGold with high potential of being targeted. No changes could be detected for any of the genes. Two biological repeats for each patient were analyzed. (C) Heatmap of amplicon sequencing result for modifications of 38 intronic off-targets predicted by CRISPOR and CrispRGold. Percentages plotted as in (A). None of the off-targets show a higher amount of modified reads in the edited vs. the unedited control. Two biological repeats from patient 2 were analyzed.