Figure 6.
Functional recovery of CAPN3 expression
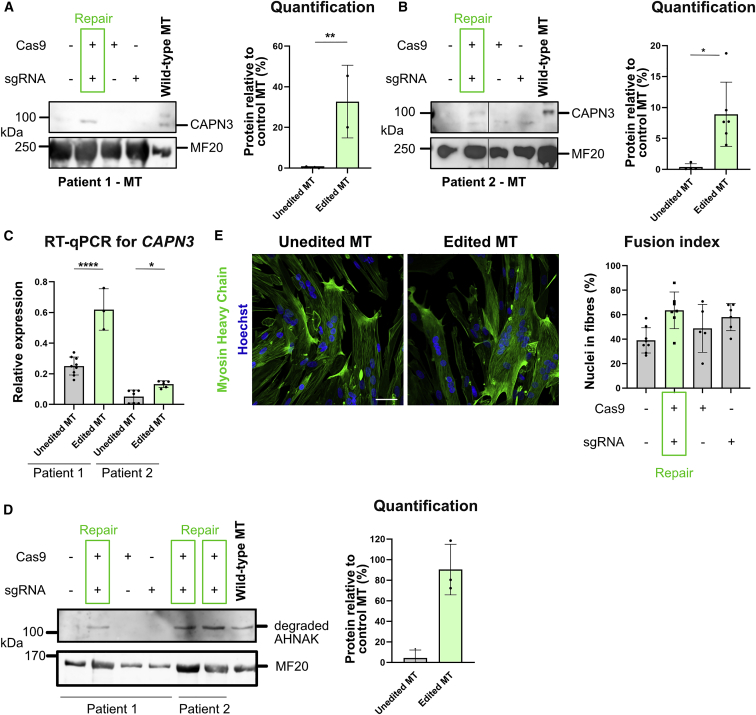
(A) Left: western blot from myotubes (MTs) for CAPN3 after gene repair in patient 1. CAPN3 can be detected in edited MT. Right: quantification. Differentiated MTs that were edited show 30% protein recovery. The intensity of the CAPN3 band was normalized to MF20. Control MT are derived from healthy controls. Statistical analysis was done using unpaired t test; p < 0.0005; n = 2. (B) As in (A) for patient 2. Left: western blot. CAPN3 can be detected after editing. Lanes were run on the same gel, but noncontinuous. Right: quantification. Edited and differentiated MTs show 10% protein recovery compared with control MT. The intensity of the CAPN3 band was normalized to MF20. Control MT are derived from healthy controls. Statistical analysis was done using unpaired t test; p < 0.05, n = 5. (C) Detection of CAPN3 expression by qRT-PCR. Consistent with the western blot results, CAPN3 expression is recovered for both patients after editing. Relative expression was calculated to GAPDH. Statistics were done using one-way analysis of variance (ANOVA); p (patient 1) < 0.0001; p (patient 2) > 0.01), n = 3. (D) Western Blot from MTs for degradation of AHNAK after gene repair in patients 1 and 2. Degradation of AHNAK can be detected in edited MT. Control MT are derived from healthy controls. (E) Unedited or edited patient PHSats were differentiated into multinucleated MTs and stained for myosin heavy chain (MyHC). Nuclei were counterstained with Hoechst and fusion index for patient 2 was determined. Scale bar, 50 μm.