Figure 17.
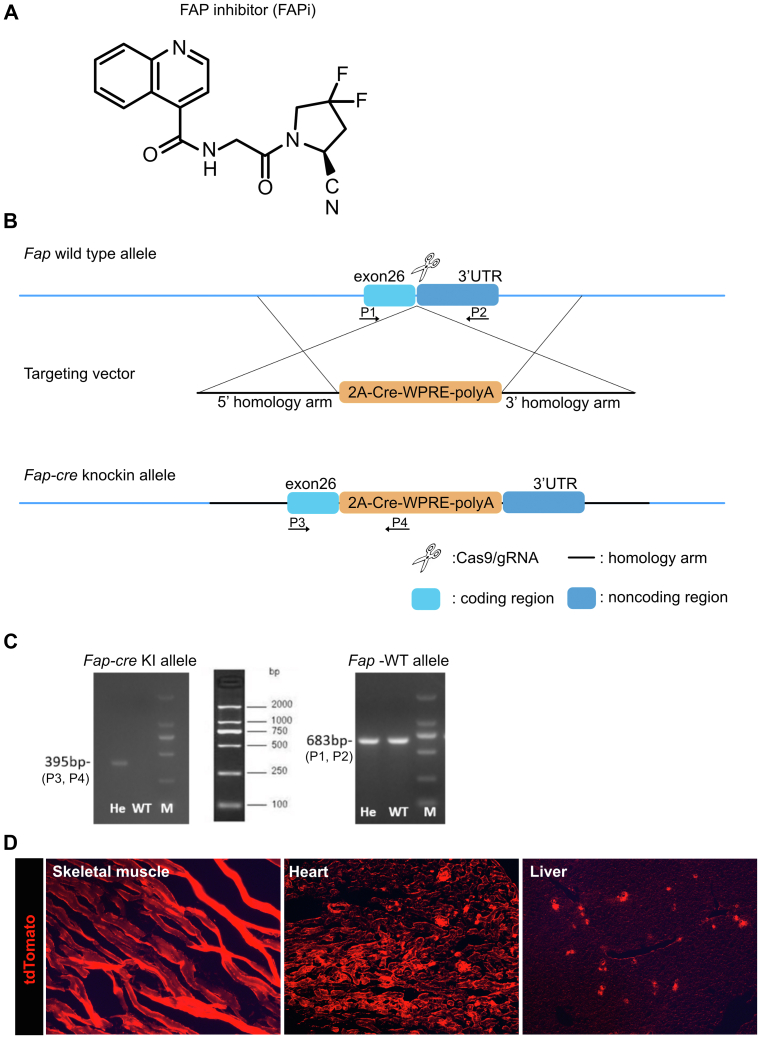
Formula and specificity of the FAP inhibitor (A) and targeting strategy to generate Fap-cre-knock in (KI) mice (B–C). (B), Diagram of the Fap-WT and Fap-Cre-KI alleles. C57BL/6 embryonic stem cells were transfected by the CRISPR/Cas9 technology using 2 ∼ 3.0 kb homologous arms on both sides of the 2A-Cre-WPRE (woodchuck posttranscriptional regulatory element)-polyA insertion sequence. Positive clones were selected with ampicillin. The Cre element in the Fap-cre-KI were inserted in exon 26, between the Fap STOP and polyadenylation signals. After embryo transfer, founder mice (F0) with successful homologous recombination were bred with C57BL/6 mice to obtain the F1 generation (Biomodel Organism Science & Technology Development, Shanghai). (C), Genotyping of WT and heterozygous (He) mice by PCR using primer pairs specific to the Fap-cre allele and Fap-WT allele confirmed correct insertion. To detect the constitutive KI allele (395-bp fragment) as well as the WT allele (683-bp fragment) by PCR. The following primers were used: (P3) 5′-CTTAATGCACCAGTTCTATC-3′, forward; (P4) 5′-GAGCATCTTCCAGGTGTG -3′, reverse; (P1) 5′-TGCCGCACTTATGCAATGAAGACAAT-3′, forward; (P2) 5′-CCCGGCAATGAACAGGTGATAAAACA-3′, reverse. 2A: ‘self-cleaving’ 2A peptide from porcine teschovirus-1. (D), tdTomato is expressed in FAP-positive cells upon crossing the reporter mice (LSL-tdTomato) with FAP-cre mice. FAP-expressing cells are detected in muscle cells of striated muscle, heart, and liver.