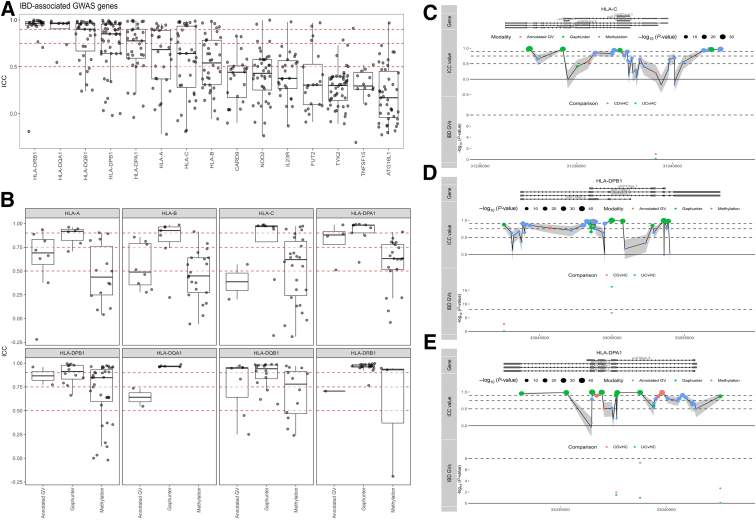
Figure 9.
IBD risk genes. (A), Visualisation of the ICCs for all CpGs annotated to IBD-associated GWAS genes. Box plots show the overall median stability within each gene. Red dashed lines represent the classification boundaries introduced by Koo and Li,47 with blocks representing poor (ICC < 0.5), moderate (0.5 ≤ ICC < 0.75), good (0.75 ≤ ICC < 0.9), and excellent (0.9 ≥ ICC). (B), ICC values of all CpG loci of class I and II HLA genes. The potential GV representing probes that were annotated with a genetic variant, predicted GV representing probes that presented a methylation signal typically found when driven by a GV, and methylation representing probes for which we have no evidence that they hybridize with any GV. Visualizations of the ICC values of all Illumina CpGs annotated to HLA-C (C), HLA-DPB1 (D), and HLA-DPA1 (E), relative to their position on each gene and grouped as potential GV (pink), predicted GV (green), or methylation (blue). Dots below represent known genetic variants as reported by Goyette53 for CD vs healthy controls (pink) and UC vs healthy controls (turquoise).