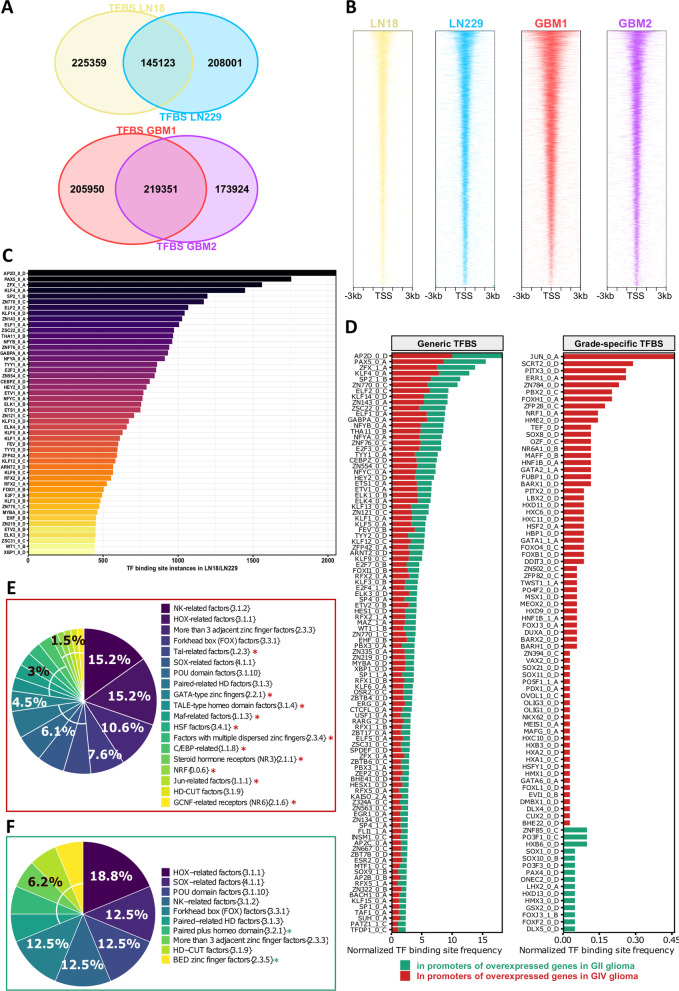
Fig. 1.
Global characterization of transcription factor binding sites in open chromatin regions in glioblastoma cell lines and glioblastoma specimens. A Total number of predicted transcription factor binding sites (TFBS) in open-chromatin regions using ATAC-seq fragments and position-weight matrices (PWMs) motifs in established human glioma cell lines LN18 and LN229 and in glioblastoma samples. In silico TFBS predictions for both cell lines were selected for downstream analysis. B Profile heatmap of total ATAC-seq peaks identified around transcription start sites (TSS) in cell lines and GBM specimens. C Top 50 occurrences of TFBS (identified with HOmo sapiens COmprehensive MOdel COllection - HOCOMOCO v11) in gene promoters (TSS ± 1.5 kb) in LN18 and LN229 glioma cell lines; the last letter (A–D) represents the quality, where A represents motifs with the highest confidence and the number defines the motif rank, with zero indicating the primary model (primary binding preferences). D Prediction of “generic” (left-panel) and “grade-specific” (right-panel) TFBS in the promoters (TSS ± 1.5 kb) of dysregulated genes in gliomas of grade IV and II. The abscissa represents a normalization factor for TFBS occurrences in which the total number of differentially expressed genes in a given glioma grade is taken into account. E, F Transcription factor (TF) families (HOCOMOCOv11) with putative binding sites in the promoters of overexpressed genes in IV glioma (E) and grade II glioma (F). Unique TF families found in either group are highlighted with asterisks