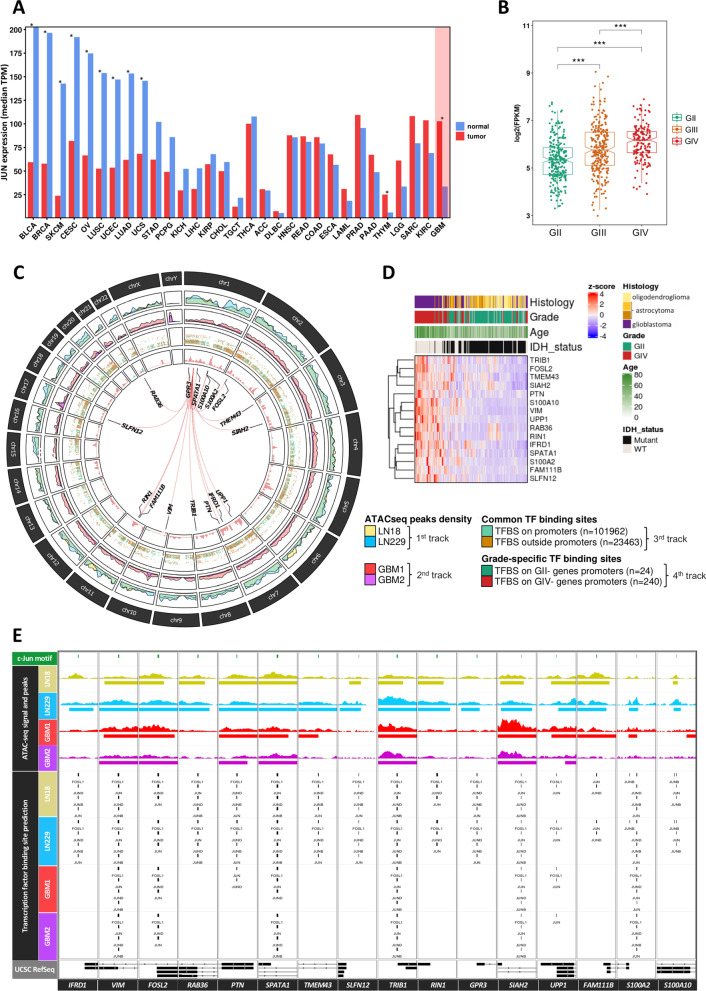
Fig. 2.
c-Jun dysregulation in the Pan-cancer atlas and the TCGA glioma dataset and identification of genomic targets in open-chromatin regions. A JUN mRNA expression profile ordered by expression differences between tumor samples (TCGA) and paired normal tissues (TCGA normal + GTEx normal). The differential expression was calculated using one-way ANOVA (Tumors or Normal, *p value < 0.05). B c-Jun mRNA expression across glioma grades using the TCGA data. The differential expression was calculated using Wilcoxon rank-sum statistical test (*p value < 0.05, **p value < 0.01 and ***p value < 0.001). C Chromatin accessibility profiling (1st and 2nd tracks) and TFBS predictions in or outside promoters (3rd track). The 4th track depicts TFBS predictions in overexpressed genes in certain glioma grades, and red lines connect JUN gene location (chr1:58,776,845–58,784,048) to each of the c-Jun-controlled genes in GBM. D Unsupervised hierarchical clustering heatmap of c-Jun gene targets in grade II and grade IV gliomas from the TCGA dataset (248 grade II gliomas; 160 grade IV gliomas). E Landscape of c-Jun binding prediction in the cis-regulatory regions of selected overexpressed GIV genes, in the studied cell lines and GBM patient samples. Location of the identified c-Jun motif is shown in green. The ATAC-seq signal and MACS2 broad peaks for each cell line and GBM patient sample are shown separately. Exons (rectangles) and introns (lines) are depicted as well as the gene orientation (arrows) in the UCSC gene composite track