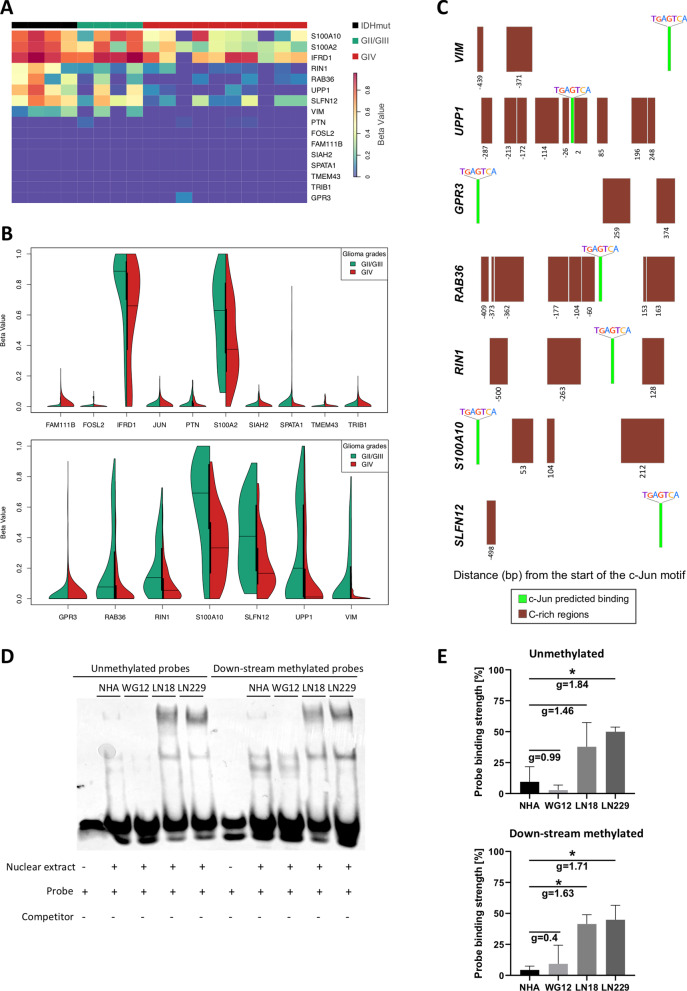
Fig. 6.
DNA methylation differs in cis-regulatory regions in c-Jun gene targets. A Heatmap of hierarchical clustering analysis showing median DNA methylation of promoters of c-Jun target genes (2 kb upstream and 500 bp downstream relative to TSS) in glioma samples. The following labels were used: IDHmut (4 GII/GIII tumors), GII/GIII (4 only GII/GIII-IDHwt tumors) and GIV (10 IDHwt tumors). DNA methylation levels showed as beta values, with 0.0–0.2 representing hypomethylated cytosines and 0.6–1.0 representing hypermethylated cytosines. B Differences in beta values distribution in gene promoters with the predicted c-Jun TFBS in GII/GIII-IDHwt versus GIV glioma samples that are statistically non-significant (upper panel) and statistically significant (bottom panel). C Distance of c-Jun motif to the beginning of differentially methylated C-rich regions between high- and low-grade gliomas. Green boxes represent a c-Jun predicted binding site, while brown boxes show each C-rich region that was found significantly differently methylated between low- and high-grade glioma samples (Chi-squared test at significance level adjusted p < 0.05). D Competitive electrophoretic mobility assay (EMSA) was used for measuring binding affinity of nuclear extracts from tumor derived cell lines and NHA, to the methylated and unmethylated double-stranded DNA from the UPP1 promoter site. Binding of proteins from NHA, WG12, LN18, and LN229 to the methylated and unmethylated probes was evaluated. Lane 1, 6: unlabeled probes; lanes 2–5 and 7–10: protein binding. E Probe binding strength in EMSA densitometry analysis is expressed as a percentage of the overall variation across all cell lines. One-way ANOVA with Fisher's LSD post hoc tests revealed significant differences at *p < 0.05 and **p < 0.01; Hedge's effect size (g) indicates the strength of the difference of the binding between the studied cell lines (medium effects g ≈ 0.5; large effects g ≈ 0.8), n = 3 ± SD