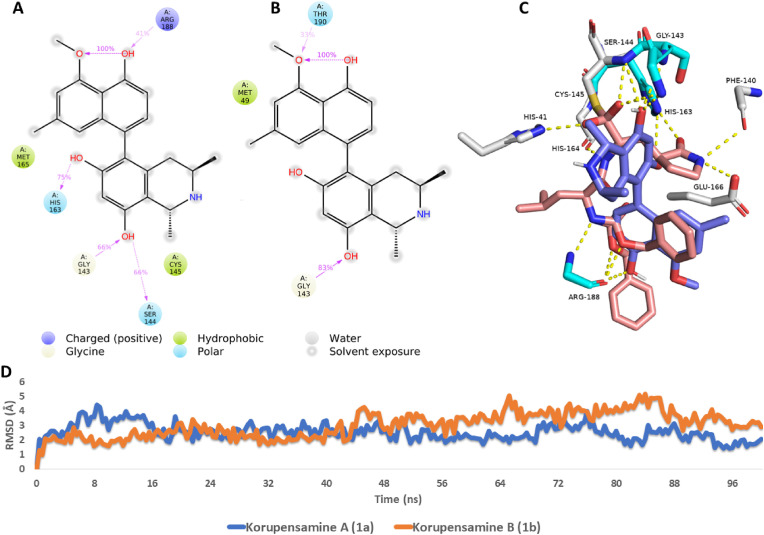
Fig. 5.
Ligand-protein interactions extracted from the 100-ns long MDS experiments of (A) korupensamine A (1a) and (B) korupensamine B (1b). 3D binding mode of korupensamine A (1a, in blue) in alignment with the reference inhibitor GC376 (brick-red structures) inside the active site of Mpro (C). The cyan-colored amino acid residues interact with both structures (i.e., korupensamine A and GC376), while white-colored amino acid residues interact with GC376 only. RMSDs of korupensamines A (1a) and B (1b) over the course of their MDS runs (D). Colored arrows in (A) and (B) indicate the percentage of interaction with each amino acid residue during the MDS runs.