Figure 7. R-spondin 2 orchestrates pathological crosstalk between joint-resident cell types.
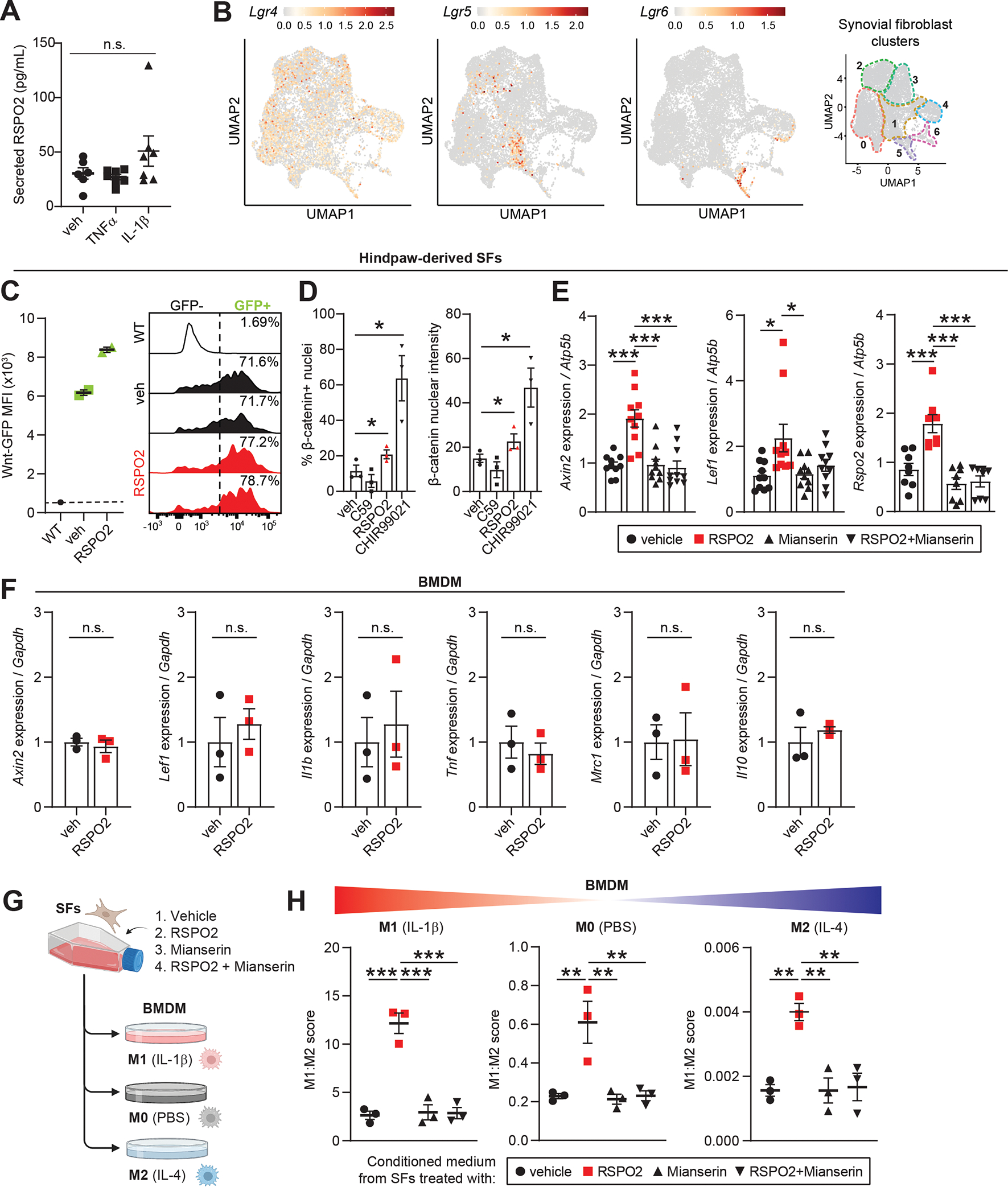
(A) Secreted RSPO2 protein levels in conditioned medium from cultured knee-derived SFs treated for 48 h with vehicle (veh), TNFα (10 ng/mL) or IL-1β (10 ng/mL). (B) Feature plots of Lgr receptors (Lgr4–6) in SFs. Color scales are not equivalent between plots. Cluster borders are shown on the right. (C) Hindpaw-derived SFs from Wnt-GFP reporter mice were treated with veh or RSPO2 (200 ng/mL) for 24 h. SFs were analyzed by flow cytometry to assess Wnt signaling activity (Wnt-GFP MFI, left) and the total percentage of Wnt-GFP+ cells (right). WT SFs (no reporter) were used as a negative control. Two biological replicates were performed. (D) Hindpaw-derived SFs were treated for 4 h with veh, C59 (0.25 μM), RSPO2 (200 ng/mL), or CHIR99021 (10 μM) on 1.5 kPa Ibidi soft substrate dishes (n=3 biological replicates). Immunocytochemical staining of non-phosphorylated/active β-catenin was performed to assess nuclear localization. Left: percentage of cells with nuclei positively stained for β-catenin. Right: Nuclear β-catenin staining intensity. (E) Hindpaw-derived SFs were treated for 24 h with veh, RSPO2 (200 ng/mL), Mianserin (20 μM), or RSPO2+Mianserin (n=8–10 biological replicates). Expression of Axin2, Lef1 and Rspo2 was normalized to Atp5b levels, and veh samples were set to 1. (F) Bone marrow-derived macrophages (BMDM) were treated for 8 h with veh or RSPO2 (200 ng/mL) (n=3 biological replicates). Expression of Axin2, Lef1, Il1b, Tnf, Mrc1 and Il10 were normalized to Gapdh levels, and veh samples were set to 1. (G) Schematic showing experimental design of SF-BMDM conditioned media treatment in (H). (H) Hindpaw-derived SFs were treated with vehicle, RPSO2 (200 ng/mL), Mianserin (20 μM), or RSPO2+Mianserin. After 6 h, treatment media was removed, cells were washed and replenished with normal SF media for another 18 h (24 h total). SF conditioned media was harvested and directly treated to BMDM for 8 h in tandem with M0 polarization (PBS, middle), M1 polarization (10 ng/mL IL-1β, left), or M2 polarization (10 ng/mL IL-4, right) (n=3 biological replicates per condition). A composite M1:M2 polarization score was calculated based on the expression of M1 (Il1b, Il6, Nos2) and M2 (Mrc1, Il10, Arg1) genes by qPCR. Gapdh was used as the housekeeper gene. For A, E and H, one-way ANOVA with multiple comparisons and Tukey’s post hoc testing was performed where *P<0.05, **P<0.01, ***P<0.001. For D and E, paired two-tailed student’s t-tests were performed where *P<0.05. Error bars are mean ± SEM. n.s.: not significant.
