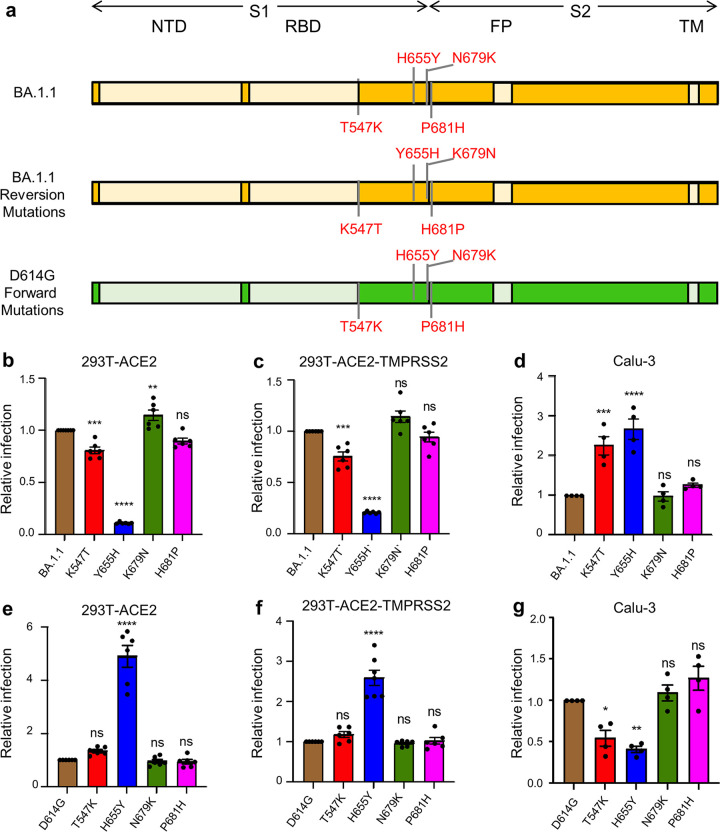
FIG 1.
H655Y governs differential entry of Omicron BA.1.1 into distinct target cells. (a) Schematic representations of BA.1.1 and D614G spike glycoprotein are presented. The N-terminal domain (NTD), the receptor binding domain (RBD), the fusion peptide (FP), and the transmembrane (TM) region are indicated. Only the mutations at the C terminus of S1 and those near the S1/S2 junction in BA.1.1 S relative to SARS-CoV-2 D614G S are shown (top, BA.1.1) (43); also displayed are these four amino acid mutations of BA.1.1 S replaced with the corresponding amino acid in the D614G S (middle, BA.1.1 reversion mutations) and these four residues in D614G S substituted with the corresponding amino acid in BA.1.1 S (bottom, D614G forward mutations). (b to d) The relative infectivity of pseudotyped viruses encoding BA.1.1 S with single mutated amino acids replaced with the corresponding amino acid in the D614G S in HEK293T-ACE2 cells (n = 6) (b), HEK293T-ACE2-TMPRSS2 cells (n = 6) (c), and Calu-3 cells (n = 4) (d). The luciferase activity of parental BA.1.1 was set as 1.0 for comparison. (e to g) The relative infectivity of pseudotyped viruses encoding D614G S with single amino acids substituted with the corresponding amino acid in the BA.1.1 S in HEK293T-ACE2 cells (n = 6) (e), HEK293T-ACE2-TMPRSS2 cells (n = 6) (f), and Calu-3 cells (n = 4) (g). The luciferase activity of parental D614G was set as 1.0. In all cases, bars represent means ± standard error, and significance was determined by one-way ANOVA with Bonferroni’s multiple testing correction. ns, P ≥ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.