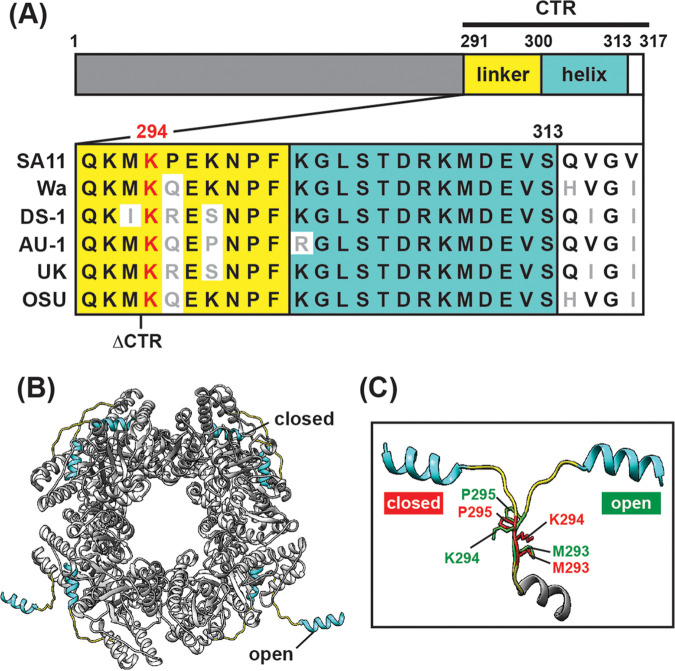
FIG 1.
Sequence conservation and structure of NSP2 CTR. (A) Cartoon schematic of NSP2 (317 amino acids in length). The core of the protein is made up of residues 1 to 290 (gray). The C-terminal region (CTR) comprises a linker (yellow; residues 291 to 300), a C-terminal α-helix (cyan; residues 301 to 313), and an unstructured region (white; residues 314 to 317). An amino acid sequence alignment of the NSP2 CTR from a variety of human and animal strains is shown below the schematic. Nonconserved residues are colored gray, and lysine 294 is colored red. The truncation location of a previously described CTR mutant (ΔCTR, missing residues 293 to 317) is indicated. (B) The octameric structure of strain SA11 NSP2 modeled in Chimera software (PDB no. 4G0A) is colored as in panel A (32). The CTR exhibits open (domain-swapping) and closed (non-domaining-swapping) conformations. (C) Zoom view of superpositioned, open and closed conformations of the NSP2 CTR. Linker residues M293, K294, and P295 are shown in stick representation and labeled (open residues are colored green, and closed residues are colored red).